Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
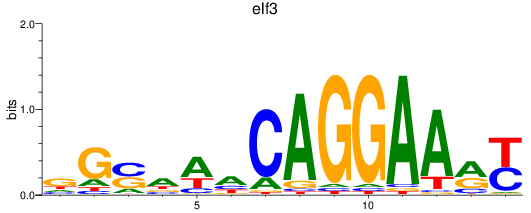
Results for elf3
Z-value: 0.82
Transcription factors associated with elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
elf3
|
ENSDARG00000077982 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| elf3 | dr11_v1_chr22_+_661711_661711 | 0.23 | 2.4e-02 | Click! |
Activity profile of elf3 motif
Sorted Z-values of elf3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_24245682 | 4.81 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr7_-_38633867 | 4.61 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr17_-_6730247 | 4.23 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr14_-_32016615 | 3.57 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr19_+_30662529 | 3.52 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr6_-_19351495 | 3.45 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr13_+_30054996 | 3.34 |
ENSDART00000110061
ENSDART00000186045 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr13_+_27316632 | 3.28 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr13_+_27316934 | 3.24 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr24_-_24146875 | 3.18 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr16_-_25393699 | 3.16 |
ENSDART00000149445
|
pard6ga
|
par-6 family cell polarity regulator gamma a |
| chr1_-_625875 | 3.12 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr5_+_32141790 | 3.00 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr4_-_27301356 | 2.92 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr12_+_30705769 | 2.90 |
ENSDART00000186448
ENSDART00000066259 |
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr10_+_3507861 | 2.87 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr10_+_21576909 | 2.87 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr22_-_16377960 | 2.84 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr20_-_22798794 | 2.76 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr17_+_19626479 | 2.76 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr25_+_34749812 | 2.75 |
ENSDART00000185712
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_31112436 | 2.65 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr16_+_14033121 | 2.61 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr23_+_44745317 | 2.60 |
ENSDART00000165654
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr4_-_12978925 | 2.59 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr22_+_2598998 | 2.57 |
ENSDART00000176665
|
CU570894.1
|
|
| chr4_-_64284924 | 2.56 |
ENSDART00000166733
|
CT956002.2
|
|
| chr13_+_31070181 | 2.55 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr3_-_50277959 | 2.55 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr13_+_30055171 | 2.38 |
ENSDART00000143581
ENSDART00000132027 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr1_-_46718353 | 2.34 |
ENSDART00000074514
|
spryd7a
|
SPRY domain containing 7a |
| chr4_-_4261673 | 2.32 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr5_+_52067723 | 2.24 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr10_+_32561317 | 2.24 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr13_-_21701323 | 2.22 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr2_+_31838442 | 2.21 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr12_-_43435254 | 2.21 |
ENSDART00000182723
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr20_-_18382708 | 2.20 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr12_-_16084835 | 2.16 |
ENSDART00000090881
|
kcnj19b
|
potassium voltage-gated channel, subfamily J, member 19b |
| chr3_+_46479705 | 2.15 |
ENSDART00000181564
|
tyk2
|
tyrosine kinase 2 |
| chr8_+_50742975 | 2.06 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr1_+_10318089 | 2.06 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr20_+_38285671 | 2.05 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr5_+_42467867 | 2.04 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr1_-_18848955 | 2.03 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr6_-_17849786 | 2.00 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr15_+_15415623 | 1.99 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr1_+_1904419 | 1.95 |
ENSDART00000142874
|
si:ch211-132g1.4
|
si:ch211-132g1.4 |
| chr7_+_48460239 | 1.95 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr1_+_51386649 | 1.93 |
ENSDART00000152289
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr16_+_1802307 | 1.88 |
ENSDART00000180026
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr10_-_41450367 | 1.86 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr15_-_2010926 | 1.86 |
ENSDART00000155688
|
dock10
|
dedicator of cytokinesis 10 |
| chr16_-_55028740 | 1.82 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr11_-_18601955 | 1.80 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr14_-_34276574 | 1.79 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr8_+_8712446 | 1.79 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr2_+_25929619 | 1.78 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr7_-_7676186 | 1.76 |
ENSDART00000091105
|
mfap3l
|
microfibril associated protein 3 like |
| chr13_+_33304187 | 1.76 |
ENSDART00000075826
ENSDART00000145295 |
dcdc2b
|
doublecortin domain containing 2B |
| chr9_+_6079528 | 1.75 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr21_-_2217685 | 1.74 |
ENSDART00000159315
|
si:dkey-50i6.5
|
si:dkey-50i6.5 |
| chr6_-_19340889 | 1.73 |
ENSDART00000181407
|
mif4gda
|
MIF4G domain containing a |
| chr17_-_44440832 | 1.72 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr16_+_34528409 | 1.71 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr6_+_32393057 | 1.70 |
ENSDART00000190765
|
dock7
|
dedicator of cytokinesis 7 |
| chr14_+_20911310 | 1.70 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr15_-_26570948 | 1.70 |
ENSDART00000156621
|
wdr81
|
WD repeat domain 81 |
| chr22_-_20342260 | 1.67 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr1_+_40237276 | 1.66 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr20_-_13625588 | 1.66 |
ENSDART00000078893
|
sytl3
|
synaptotagmin-like 3 |
| chr16_-_16590780 | 1.66 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr12_+_30706158 | 1.62 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr9_-_25425381 | 1.62 |
ENSDART00000129522
|
acvr2aa
|
activin A receptor type 2Aa |
| chr11_-_29816429 | 1.60 |
ENSDART00000069132
|
cybb
|
cytochrome b-245, beta polypeptide (chronic granulomatous disease) |
| chr16_+_46807214 | 1.60 |
ENSDART00000185524
|
CABZ01027551.1
|
|
| chr3_-_30625219 | 1.60 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr8_-_11202378 | 1.59 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr14_+_28438947 | 1.58 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr15_+_16525126 | 1.58 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr2_-_59327299 | 1.54 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr2_+_27652386 | 1.54 |
ENSDART00000188261
|
tmem68
|
transmembrane protein 68 |
| chr22_-_21176269 | 1.54 |
ENSDART00000112839
|
rex1bd
|
required for excision 1-B domain containing |
| chr23_+_42304602 | 1.53 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr24_-_9960290 | 1.52 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr4_+_2655358 | 1.52 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr1_+_55293424 | 1.51 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr22_-_16377666 | 1.50 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr13_+_30696286 | 1.49 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr18_-_48508585 | 1.49 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr5_-_35137365 | 1.48 |
ENSDART00000141218
|
fcho2
|
FCH domain only 2 |
| chr4_-_23963838 | 1.47 |
ENSDART00000133433
ENSDART00000132615 ENSDART00000135942 ENSDART00000139439 |
celf2
|
cugbp, Elav-like family member 2 |
| chr3_-_24205339 | 1.47 |
ENSDART00000157135
|
si:dkey-110g7.8
|
si:dkey-110g7.8 |
| chr17_-_51223788 | 1.47 |
ENSDART00000149864
|
psen1
|
presenilin 1 |
| chr5_-_37959874 | 1.47 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr6_-_41079209 | 1.46 |
ENSDART00000151592
|
rab44
|
RAB44, member RAS oncogene family |
| chr19_-_20430892 | 1.44 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr4_+_26629368 | 1.43 |
ENSDART00000183575
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr3_-_37571601 | 1.43 |
ENSDART00000016407
|
arf2a
|
ADP-ribosylation factor 2a |
| chr2_+_36721357 | 1.43 |
ENSDART00000019063
|
pdcd10b
|
programmed cell death 10b |
| chr9_-_49860756 | 1.42 |
ENSDART00000044270
|
ttc21b
|
tetratricopeptide repeat domain 21B |
| chr19_+_42693855 | 1.41 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr1_+_36722122 | 1.41 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr3_+_57997980 | 1.40 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr10_-_22828302 | 1.40 |
ENSDART00000079459
ENSDART00000100468 |
per1a
|
period circadian clock 1a |
| chr7_+_16509201 | 1.40 |
ENSDART00000173777
|
zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr11_+_44226200 | 1.39 |
ENSDART00000191417
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr6_+_37625787 | 1.38 |
ENSDART00000065122
|
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr24_-_22756508 | 1.37 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr10_+_1668106 | 1.36 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr3_-_34337969 | 1.35 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr1_+_27867706 | 1.33 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr20_+_27087539 | 1.33 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr12_-_33558879 | 1.33 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr14_+_14043793 | 1.32 |
ENSDART00000164376
|
rraga
|
Ras-related GTP binding A |
| chr22_+_2300484 | 1.32 |
ENSDART00000106503
|
znf1180
|
zinc finger protein 1180 |
| chr8_-_52715911 | 1.32 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr21_-_40173821 | 1.32 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr16_-_16590489 | 1.31 |
ENSDART00000190021
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr24_+_35911020 | 1.31 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr7_-_24204665 | 1.30 |
ENSDART00000167141
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr3_+_46479913 | 1.30 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr5_-_45894802 | 1.30 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr2_-_2096055 | 1.28 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr5_-_55623443 | 1.27 |
ENSDART00000005671
ENSDART00000176341 |
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr23_-_26120281 | 1.27 |
ENSDART00000133372
ENSDART00000103872 |
rca2.1
|
regulator of complement activation group 2 gene 1 |
| chr4_+_4902392 | 1.26 |
ENSDART00000133866
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr18_+_11970987 | 1.25 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr17_-_19626357 | 1.24 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr21_-_14762944 | 1.24 |
ENSDART00000114096
|
arrdc1b
|
arrestin domain containing 1b |
| chr12_+_33395748 | 1.24 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr2_-_30200206 | 1.24 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr22_-_16403602 | 1.23 |
ENSDART00000183417
|
lama3
|
laminin, alpha 3 |
| chr2_+_27651984 | 1.23 |
ENSDART00000126762
|
tmem68
|
transmembrane protein 68 |
| chr24_-_35707552 | 1.22 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr22_+_1586060 | 1.21 |
ENSDART00000160793
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr19_-_24125457 | 1.21 |
ENSDART00000080632
|
zgc:64022
|
zgc:64022 |
| chr12_-_33558727 | 1.21 |
ENSDART00000086087
|
mbtd1
|
mbt domain containing 1 |
| chr22_+_12798569 | 1.21 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr22_-_16758438 | 1.21 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr1_-_54997746 | 1.20 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr23_-_10175898 | 1.19 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr9_+_48085039 | 1.19 |
ENSDART00000186734
|
si:ch73-54b5.2
|
si:ch73-54b5.2 |
| chr8_-_52594111 | 1.18 |
ENSDART00000167667
|
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr7_+_41748693 | 1.18 |
ENSDART00000174379
ENSDART00000052168 |
hrh3
|
histamine receptor H3 |
| chr21_+_35296246 | 1.18 |
ENSDART00000076750
|
il12bb
|
interleukin 12B, b |
| chr22_+_1734981 | 1.18 |
ENSDART00000158195
|
znf1159
|
zinc finger protein 1159 |
| chr10_-_26766780 | 1.18 |
ENSDART00000146666
|
mcf2b
|
MCF.2 cell line derived transforming sequence b |
| chr10_+_10636237 | 1.16 |
ENSDART00000136853
|
fam163b
|
family with sequence similarity 163, member B |
| chr3_-_32902138 | 1.15 |
ENSDART00000144026
ENSDART00000083874 ENSDART00000145443 ENSDART00000148239 ENSDART00000134917 |
kat7a
|
K(lysine) acetyltransferase 7a |
| chr6_-_19341184 | 1.15 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr5_-_41124241 | 1.14 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr4_-_17642168 | 1.14 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr17_-_36988455 | 1.14 |
ENSDART00000187180
ENSDART00000126823 |
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr24_-_11127493 | 1.14 |
ENSDART00000144066
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr13_+_4225173 | 1.14 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr3_+_46762703 | 1.13 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr2_+_36109002 | 1.12 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr13_+_28705143 | 1.11 |
ENSDART00000183338
|
ldb1a
|
LIM domain binding 1a |
| chr7_+_31051603 | 1.11 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr20_-_51727860 | 1.11 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr11_+_25139495 | 1.11 |
ENSDART00000168368
|
si:ch211-25d12.7
|
si:ch211-25d12.7 |
| chr4_-_77252368 | 1.11 |
ENSDART00000111941
|
zgc:174310
|
zgc:174310 |
| chr17_+_43468732 | 1.10 |
ENSDART00000055487
|
chmp3
|
charged multivesicular body protein 3 |
| chr7_-_51476276 | 1.10 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr20_-_1191910 | 1.10 |
ENSDART00000043218
|
ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr14_+_26796684 | 1.09 |
ENSDART00000187414
|
klhl4
|
kelch-like family member 4 |
| chr23_-_42810664 | 1.09 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr15_-_2652640 | 1.08 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr1_+_12767318 | 1.08 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr11_+_20899029 | 1.06 |
ENSDART00000163029
|
zgc:162182
|
zgc:162182 |
| chr8_+_20157798 | 1.06 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr6_+_19689464 | 1.05 |
ENSDART00000164960
|
stx8
|
syntaxin 8 |
| chr13_+_34689663 | 1.05 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr2_+_3823813 | 1.05 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr12_-_44043285 | 1.05 |
ENSDART00000163074
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr10_+_39304422 | 1.04 |
ENSDART00000019267
|
kcnj1b
|
potassium inwardly-rectifying channel, subfamily J, member 1b |
| chr4_-_30422325 | 1.03 |
ENSDART00000158444
|
znf1114
|
zinc finger protein 1114 |
| chr11_+_38280454 | 1.03 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr23_+_32021803 | 1.02 |
ENSDART00000012963
|
trappc6b
|
trafficking protein particle complex 6b |
| chr1_-_14258409 | 1.02 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr2_-_36918709 | 1.02 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr7_-_6351021 | 1.02 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr12_+_14556092 | 1.02 |
ENSDART00000115237
|
becn1
|
beclin 1, autophagy related |
| chr10_-_105100 | 1.02 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr10_+_42169982 | 1.01 |
ENSDART00000190905
|
CU467905.1
|
|
| chr19_-_12404590 | 1.01 |
ENSDART00000103703
|
ftr56
|
finTRIM family, member 56 |
| chr6_-_28943056 | 1.01 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr1_+_1896737 | 1.00 |
ENSDART00000152442
|
si:ch211-132g1.6
|
si:ch211-132g1.6 |
| chr15_+_3808996 | 1.00 |
ENSDART00000110227
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr16_-_22251414 | 1.00 |
ENSDART00000158500
ENSDART00000179998 |
atp8b2
|
ATPase phospholipid transporting 8B2 |
| chr4_-_22310956 | 0.99 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr3_-_21061931 | 0.99 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr8_-_39838660 | 0.99 |
ENSDART00000139266
|
ftr98
|
finTRIM family, member 98 |
| chr5_-_61797220 | 0.98 |
ENSDART00000079855
|
im:7138535
|
im:7138535 |
| chr8_-_36399884 | 0.98 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr6_-_44161262 | 0.97 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr12_-_23009312 | 0.97 |
ENSDART00000111801
|
mkxa
|
mohawk homeobox a |
| chr16_+_43368572 | 0.96 |
ENSDART00000032778
ENSDART00000193897 |
rnf144b
|
ring finger protein 144B |
| chr7_+_60161002 | 0.96 |
ENSDART00000054007
|
slc8a4b
|
solute carrier family 8 (sodium/calcium exchanger), member 4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of elf3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.6 | 1.7 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.5 | 1.4 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.5 | 2.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 1.3 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.4 | 7.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.4 | 1.5 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.4 | 1.4 | GO:2000383 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.3 | 1.0 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.3 | 1.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.3 | 2.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 2.8 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 0.8 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.3 | 1.6 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.2 | 0.7 | GO:0042832 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.2 | 0.7 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.2 | 1.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 6.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 1.0 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 0.8 | GO:0045938 | regulation of endocrine process(GO:0044060) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) endocrine hormone secretion(GO:0060986) |
| 0.2 | 4.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 4.0 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 1.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 0.5 | GO:0002902 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.2 | 1.9 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 1.4 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.6 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 1.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 2.8 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.6 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.6 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 1.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.1 | 0.5 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 1.0 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 1.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 6.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.2 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.7 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 5.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.9 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.8 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 0.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.9 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.4 | GO:0042542 | response to hydrogen peroxide(GO:0042542) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 3.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.1 | 0.2 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 1.3 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 1.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.0 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 0.4 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 1.7 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 1.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 2.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 2.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.6 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.1 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.9 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.8 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.0 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 1.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 2.0 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 1.0 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 1.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.7 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 1.5 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 2.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.1 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.7 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.3 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 1.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 2.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 2.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.2 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 4.1 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.1 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 2.8 | GO:0061008 | hepaticobiliary system development(GO:0061008) |
| 0.0 | 1.1 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.6 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 1.1 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 3.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.3 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 1.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.2 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 1.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 3.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.4 | 1.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.4 | 1.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.4 | 2.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 1.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.3 | 1.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.3 | 2.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.3 | 1.2 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.3 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 4.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 1.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 1.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 1.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 2.6 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 3.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 1.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 2.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.6 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 5.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.8 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 3.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 5.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 2.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 5.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 3.8 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.9 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 3.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.4 | 4.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 1.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.4 | 1.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 4.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.3 | 1.2 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.3 | 0.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 1.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 2.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 2.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 0.7 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.2 | 1.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.8 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.2 | 1.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 0.9 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 1.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 2.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.8 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 1.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.5 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 1.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 0.7 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 2.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 6.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.4 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.5 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.4 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 5.1 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 2.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 2.8 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 1.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 2.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 1.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 1.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.9 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.1 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 6.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.6 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 17.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 9.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 4.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 2.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.3 | 2.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |