Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for en1b_en2a+en2b_gbx1_emx1
Z-value: 0.99
Transcription factors associated with en1b_en2a+en2b_gbx1_emx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
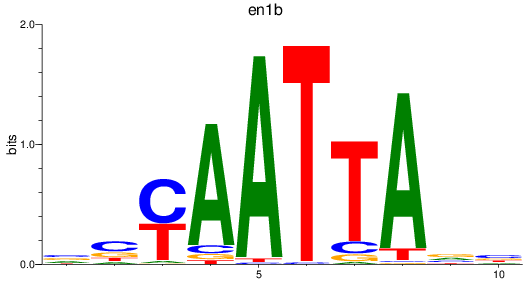
en1b
|
ENSDARG00000098730 | engrailed homeobox 1b |
|
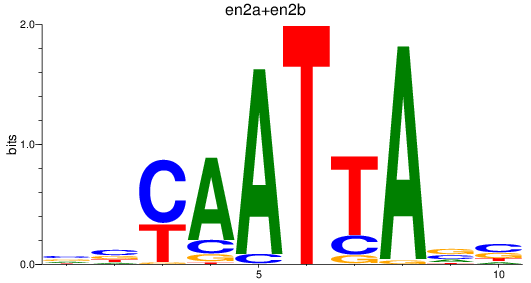
en2a
|
ENSDARG00000026599 | engrailed homeobox 2a |
|
en2b
|
ENSDARG00000038868 | engrailed homeobox 2b |
|
en2a
|
ENSDARG00000115233 | engrailed homeobox 2a |
|
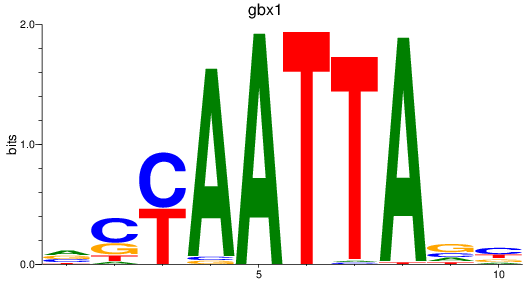
gbx1
|
ENSDARG00000071418 | gastrulation brain homeobox 1 |
|
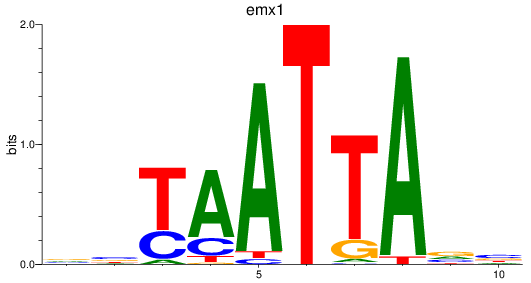
emx1
|
ENSDARG00000039569 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en2a | dr11_v1_chr7_-_40993456_40993456 | 0.77 | 1.7e-19 | Click! |
| en1b | dr11_v1_chr1_+_7679328_7679328 | 0.76 | 1.3e-18 | Click! |
| en2b | dr11_v1_chr2_+_29976419_29976419 | 0.74 | 1.1e-17 | Click! |
| gbx1 | dr11_v1_chr24_+_34113424_34113424 | 0.70 | 6.6e-15 | Click! |
| emx1 | dr11_v1_chr13_+_15004398_15004398 | 0.22 | 3.2e-02 | Click! |
Activity profile of en1b_en2a+en2b_gbx1_emx1 motif
Sorted Z-values of en1b_en2a+en2b_gbx1_emx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_64732036 | 20.43 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr5_+_64732270 | 17.24 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr7_+_30787903 | 16.99 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr23_+_44741500 | 15.99 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr22_+_17828267 | 15.47 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr23_+_40460333 | 15.21 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr1_-_50859053 | 14.30 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr24_-_41320037 | 13.31 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr19_-_5805923 | 13.27 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr16_+_5774977 | 12.38 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr2_+_50608099 | 12.16 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr22_+_17205608 | 11.57 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr25_+_21324588 | 11.31 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr21_+_28958471 | 10.78 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_+_32722227 | 10.15 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr7_+_13418812 | 10.09 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr16_+_47207691 | 10.07 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr12_+_24342303 | 9.75 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr15_-_9272328 | 9.68 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr18_+_11506561 | 9.68 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr8_-_50888806 | 9.37 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr4_+_11384891 | 9.33 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr6_-_6487876 | 9.27 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr9_+_38372216 | 9.26 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr21_-_39177564 | 9.23 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr1_+_16397063 | 9.16 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr18_+_9615147 | 9.02 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr6_+_39232245 | 8.89 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr19_+_14921000 | 8.76 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr24_+_16547035 | 8.63 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr22_+_4488454 | 8.58 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr23_+_26946744 | 8.37 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr7_-_72261721 | 8.20 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr7_+_23515966 | 8.03 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr25_-_32869794 | 7.95 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr2_-_31936966 | 7.93 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr21_+_26390549 | 7.91 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr1_-_10647484 | 7.43 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr1_+_34696503 | 7.40 |
ENSDART00000186106
|
CR339054.2
|
|
| chr10_+_15255198 | 7.26 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr5_-_23280098 | 7.18 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr20_-_9462433 | 7.07 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr2_-_14390627 | 7.07 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr25_+_3326885 | 7.07 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr7_-_25895189 | 7.06 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr11_-_42554290 | 7.04 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr23_+_28582865 | 7.01 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr6_+_21001264 | 6.91 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr2_-_11512819 | 6.84 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr10_+_15777064 | 6.49 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr7_+_15872357 | 6.43 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr4_+_21129752 | 6.24 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr19_-_5103141 | 6.23 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_+_13637383 | 6.10 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr16_-_29437373 | 6.10 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr2_-_32558795 | 6.09 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr16_+_46111849 | 5.96 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr7_-_24699985 | 5.95 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr1_+_49266886 | 5.87 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr19_+_5543072 | 5.87 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr20_-_9436521 | 5.86 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr17_+_24722646 | 5.79 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr1_-_10647307 | 5.76 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr21_+_6751760 | 5.75 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr23_-_333457 | 5.72 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr19_-_20113696 | 5.70 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr17_+_31185276 | 5.62 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr11_+_25129013 | 5.58 |
ENSDART00000132879
|
ndrg3a
|
ndrg family member 3a |
| chr20_-_46362606 | 5.56 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr9_+_17983463 | 5.53 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr19_-_30524952 | 5.43 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr17_-_12336987 | 5.34 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr22_+_5176255 | 5.33 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr11_+_30057762 | 5.33 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr3_+_34919810 | 5.29 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr20_+_27020201 | 5.25 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr21_+_6751405 | 5.19 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr16_+_20161805 | 5.08 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr18_-_1185772 | 5.06 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr9_-_20372977 | 5.02 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr1_+_33668236 | 4.99 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr11_-_44801968 | 4.98 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr11_-_30158191 | 4.91 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr5_+_30624183 | 4.91 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr14_+_8940326 | 4.85 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr15_+_44366556 | 4.82 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr3_+_33341640 | 4.79 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr24_-_31843173 | 4.69 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr11_+_38280454 | 4.67 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr20_-_31808779 | 4.64 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr13_-_29420885 | 4.61 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr15_-_11683529 | 4.59 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr20_+_34933183 | 4.55 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr18_+_28106139 | 4.49 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr2_+_20332044 | 4.48 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr1_-_38756870 | 4.45 |
ENSDART00000130324
ENSDART00000148404 |
gpm6ab
|
glycoprotein M6Ab |
| chr15_-_33925851 | 4.44 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr21_+_34088110 | 4.44 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr1_-_47071979 | 4.43 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr7_+_71547747 | 4.42 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr11_+_28476298 | 4.42 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr19_-_31402429 | 4.39 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr5_-_12219572 | 4.39 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr15_+_5277761 | 4.37 |
ENSDART00000153954
|
si:ch1073-166e24.4
|
si:ch1073-166e24.4 |
| chr16_-_29458806 | 4.30 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr25_+_7784582 | 4.25 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr1_-_46981134 | 4.23 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr12_-_18393408 | 4.21 |
ENSDART00000159674
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr18_-_8857137 | 4.20 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr12_+_2446837 | 4.19 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr8_+_49778486 | 4.12 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr13_+_38430466 | 4.12 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr9_+_52492639 | 4.12 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr20_+_19238382 | 4.10 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr10_+_15255012 | 4.07 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr18_-_40708537 | 4.06 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr15_+_23799461 | 4.05 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr21_-_42100471 | 4.03 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_+_4849789 | 4.03 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr15_-_22074315 | 3.95 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr8_+_22930627 | 3.93 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr17_+_23298928 | 3.91 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr20_+_35382482 | 3.89 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr3_+_1637358 | 3.85 |
ENSDART00000180266
|
CR394546.5
|
|
| chr3_-_60827402 | 3.84 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr17_-_28198099 | 3.84 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr4_+_9400012 | 3.80 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr23_+_4646194 | 3.79 |
ENSDART00000092344
|
LO017700.1
|
|
| chr10_+_42690374 | 3.79 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr17_-_200316 | 3.77 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr12_+_31744217 | 3.71 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr19_-_5865766 | 3.69 |
ENSDART00000191007
|
LO018585.1
|
|
| chr23_-_14990865 | 3.68 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr13_+_15838151 | 3.65 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr9_-_32753535 | 3.64 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr5_-_21970881 | 3.63 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr14_-_32016615 | 3.63 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr2_-_11258547 | 3.62 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr14_-_34044369 | 3.60 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_+_24921587 | 3.60 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr6_-_2133737 | 3.60 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr6_+_27667359 | 3.57 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr18_+_10884996 | 3.57 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr22_+_5176693 | 3.56 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr10_+_8680730 | 3.52 |
ENSDART00000011987
|
isl1l
|
islet1, like |
| chr9_-_44295071 | 3.51 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr15_+_15856178 | 3.49 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr8_-_49908978 | 3.49 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr20_+_26349002 | 3.46 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr2_-_31302615 | 3.43 |
ENSDART00000034784
ENSDART00000060812 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr24_-_5786759 | 3.41 |
ENSDART00000152069
|
chst2b
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2b |
| chr5_-_50992690 | 3.38 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr20_-_35040041 | 3.38 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr20_-_29864390 | 3.35 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr6_-_12275836 | 3.35 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr20_+_27087539 | 3.34 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr25_+_3327071 | 3.34 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr9_-_31278048 | 3.32 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr9_-_38156894 | 3.31 |
ENSDART00000134759
|
si:dkey-219c10.4
|
si:dkey-219c10.4 |
| chr7_+_34487833 | 3.30 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr3_+_22035863 | 3.29 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr18_-_2433011 | 3.28 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr8_+_7756893 | 3.27 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr3_-_61099004 | 3.24 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr6_-_36198052 | 3.24 |
ENSDART00000086627
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr17_-_12389259 | 3.22 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr2_-_21082695 | 3.21 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr10_+_7709724 | 3.19 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr2_+_6253246 | 3.18 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr8_+_18624658 | 3.16 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr25_-_19224298 | 3.15 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr3_-_15210491 | 3.15 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr12_+_21298317 | 3.14 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr11_-_472547 | 3.13 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr15_-_590787 | 3.11 |
ENSDART00000189367
|
si:ch73-144d13.5
|
si:ch73-144d13.5 |
| chr20_-_28404362 | 3.11 |
ENSDART00000055932
ENSDART00000188161 |
pigh
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr1_+_7517454 | 3.10 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr22_+_27090136 | 3.09 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr2_+_26237322 | 3.08 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr18_+_1154189 | 3.08 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr25_-_13363286 | 3.05 |
ENSDART00000163735
ENSDART00000169119 |
ndrg4
|
NDRG family member 4 |
| chr19_-_30510259 | 3.04 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr21_+_42226113 | 3.03 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr24_-_11057305 | 3.03 |
ENSDART00000186494
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr1_-_18811517 | 3.03 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr2_-_56385373 | 3.02 |
ENSDART00000169101
|
cers4b
|
ceramide synthase 4b |
| chr1_+_18811679 | 2.98 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr15_+_33865312 | 2.98 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr6_-_48473694 | 2.96 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr6_-_40352215 | 2.92 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr5_+_37903790 | 2.92 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr3_+_13842554 | 2.92 |
ENSDART00000162317
ENSDART00000158068 |
ilf3b
|
interleukin enhancer binding factor 3b |
| chr3_+_32443395 | 2.91 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr24_-_27400017 | 2.90 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr16_-_27628994 | 2.88 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr14_-_30474346 | 2.88 |
ENSDART00000173051
ENSDART00000173124 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr13_-_36911118 | 2.87 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr4_-_15420452 | 2.86 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr24_+_10310577 | 2.86 |
ENSDART00000141718
|
otulina
|
OTU deubiquitinase with linear linkage specificity a |
| chr10_-_5581487 | 2.85 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of en1b_en2a+en2b_gbx1_emx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 37.7 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 2.5 | 10.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 2.2 | 8.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 2.1 | 6.2 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 2.0 | 7.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.8 | 18.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 1.8 | 5.3 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 1.7 | 5.0 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 1.6 | 6.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.5 | 4.6 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.5 | 6.0 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 1.5 | 4.4 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 1.4 | 5.7 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.3 | 5.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 1.3 | 6.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.2 | 5.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 1.2 | 4.7 | GO:0015677 | copper ion import(GO:0015677) |
| 1.2 | 9.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 1.1 | 3.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.0 | 4.9 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 1.0 | 2.9 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 1.0 | 2.9 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.9 | 6.4 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.9 | 4.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.9 | 3.6 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.9 | 3.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.9 | 3.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.8 | 16.0 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.8 | 2.4 | GO:0009750 | response to fructose(GO:0009750) |
| 0.8 | 11.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.8 | 4.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.7 | 2.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.7 | 3.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.7 | 10.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.7 | 2.1 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.7 | 9.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.7 | 10.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 6.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.7 | 2.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.7 | 2.0 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.6 | 1.9 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.6 | 3.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.6 | 10.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.6 | 11.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.6 | 1.7 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.6 | 2.8 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.5 | 3.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.5 | 4.8 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.5 | 2.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 2.5 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.5 | 2.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.5 | 3.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.5 | 1.9 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.4 | 4.9 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.4 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 3.4 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.4 | 12.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.4 | 1.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.4 | 7.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 12.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 5.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 | 9.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.4 | 2.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 9.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.4 | 1.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.4 | 7.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 4.4 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.3 | 1.0 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.3 | 6.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.0 | GO:0021527 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) spinal cord association neuron differentiation(GO:0021527) |
| 0.3 | 3.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.3 | 3.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.3 | 4.3 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.3 | 1.6 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.3 | 1.5 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 3.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 0.9 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.3 | 12.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 3.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.3 | 4.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.3 | 5.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.3 | 5.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.3 | 1.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.3 | 1.4 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.3 | 2.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 3.3 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.3 | 7.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.3 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 0.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.3 | 2.9 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 1.6 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.3 | 19.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.3 | 1.0 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.3 | 4.6 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.3 | 2.0 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.3 | 4.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 1.0 | GO:0048739 | response to muscle stretch(GO:0035994) cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 12.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.2 | 4.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 3.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 1.4 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 0.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 5.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 3.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.2 | 0.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 1.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 8.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 3.8 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.2 | 1.0 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 3.7 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.2 | 0.6 | GO:0045830 | regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.2 | 0.7 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 3.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 2.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.2 | 7.9 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.2 | 0.9 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 18.8 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.2 | 0.3 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) |
| 0.2 | 0.5 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 1.5 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 1.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 1.5 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.2 | 1.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 0.8 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 1.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.2 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 0.5 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.2 | 0.8 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 0.9 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 1.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 6.2 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.9 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.6 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.4 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 1.4 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.1 | 0.7 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 11.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 9.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.3 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 8.2 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 2.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.4 | GO:0042044 | fluid transport(GO:0042044) |
| 0.1 | 1.3 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.1 | 1.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.2 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.6 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 9.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.8 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.1 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 3.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 4.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 1.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 4.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.7 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 1.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 3.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.9 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 2.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.5 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 35.5 | GO:0099537 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) trans-synaptic signaling(GO:0099537) |
| 0.1 | 0.7 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 2.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.5 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 1.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 3.8 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 13.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 3.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 4.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.8 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.3 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 3.8 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.1 | 3.9 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 2.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 1.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 3.1 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.1 | 0.6 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.3 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.6 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.2 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 3.8 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.1 | 1.5 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.4 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 0.3 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 2.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 4.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 2.9 | GO:0034341 | monocyte chemotaxis(GO:0002548) response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 3.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.2 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.5 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 0.2 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.1 | 2.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.2 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.7 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.1 | 1.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 4.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.6 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 2.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 10.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.0 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 2.6 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 1.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.7 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 1.8 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 2.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 2.2 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.6 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 1.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 4.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 3.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.0 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:2000243 | positive regulation of oocyte development(GO:0060282) positive regulation of reproductive process(GO:2000243) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 1.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 2.1 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.6 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.5 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.8 | GO:0006497 | protein lipidation(GO:0006497) lipoprotein biosynthetic process(GO:0042158) |
| 0.0 | 1.2 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 1.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0033619 | membrane protein proteolysis(GO:0033619) |
| 0.0 | 1.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 1.0 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.8 | GO:0048017 | inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.2 | GO:0032324 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.9 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 1.2 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 4.8 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.8 | 18.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 1.8 | 10.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.6 | 16.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 1.2 | 6.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.2 | 7.0 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 1.1 | 5.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.1 | 6.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.0 | 11.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.9 | 9.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.8 | 13.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 11.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.8 | 10.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.6 | 3.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.6 | 10.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.6 | 2.2 | GO:0035060 | brahma complex(GO:0035060) |
| 0.5 | 1.9 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.5 | 2.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 3.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.4 | 2.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 2.9 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 12.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.3 | 12.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 1.3 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.3 | 1.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.3 | 2.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 4.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 32.0 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.3 | 5.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 3.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 5.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 7.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 2.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 4.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 13.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 5.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 1.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 3.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 4.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 13.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 31.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.2 | 4.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 3.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 4.6 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.2 | 32.1 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 85.6 | GO:0045202 | synapse(GO:0045202) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 0.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.7 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 1.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 2.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 6.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 5.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.7 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 3.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.5 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 5.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.3 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 17.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 5.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 4.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.5 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 6.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.8 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 19.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 2.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 1.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 3.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 3.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.0 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0072379 | ER membrane insertion complex(GO:0072379) |
| 0.0 | 23.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 8.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 2.8 | 11.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 2.1 | 10.5 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 2.1 | 6.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 2.0 | 7.9 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 1.8 | 10.8 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.6 | 6.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.4 | 7.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.4 | 9.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.2 | 3.7 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 1.2 | 4.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.2 | 1.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 1.1 | 10.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.0 | 18.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 1.0 | 23.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.9 | 3.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.9 | 9.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.8 | 9.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.8 | 3.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.8 | 11.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.8 | 15.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.7 | 3.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.7 | 2.0 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.7 | 5.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.7 | 2.0 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.6 | 2.6 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.6 | 4.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.6 | 1.8 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.6 | 22.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 3.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.5 | 6.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.5 | 16.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 2.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.5 | 7.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 4.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 16.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 3.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.4 | 4.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 5.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 7.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 5.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 5.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.4 | 2.6 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.4 | 2.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 13.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.4 | 1.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 2.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 1.4 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.3 | 2.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 4.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 1.6 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 11.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 8.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 0.9 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.3 | 0.9 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.3 | 1.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.3 | 3.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 4.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 7.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 1.0 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.3 | 0.8 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 3.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 4.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 7.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 1.2 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 3.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 0.9 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.2 | 2.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 7.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 3.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 6.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 5.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 8.9 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 1.0 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.2 | 1.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 4.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 3.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 0.9 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 3.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 14.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 4.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 2.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 8.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 3.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 4.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 0.5 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.2 | 8.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 0.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 1.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 0.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 3.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.2 | 2.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 1.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 5.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 3.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 2.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.0 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 1.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.6 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 7.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.9 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.8 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 1.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 2.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 3.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 2.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.0 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 2.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 4.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 4.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 0.3 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 9.7 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.1 | 0.4 | GO:0044389 | ubiquitin-like protein ligase binding(GO:0044389) |
| 0.1 | 1.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.2 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 2.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 2.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 2.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.1 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 2.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 4.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.1 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 4.9 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 13.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 11.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.5 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 3.6 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 4.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.1 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.6 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.5 | 11.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 8.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 11.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.3 | 9.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.2 | 12.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 5.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 1.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 4.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 13.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.9 | 8.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 3.8 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.6 | 10.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 8.9 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.4 | 11.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.4 | 7.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 2.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 3.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 4.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 2.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 2.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 3.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 7.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 1.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 3.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.8 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 3.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.6 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 5.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 3.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME MEIOSIS | Genes involved in Meiosis |