Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
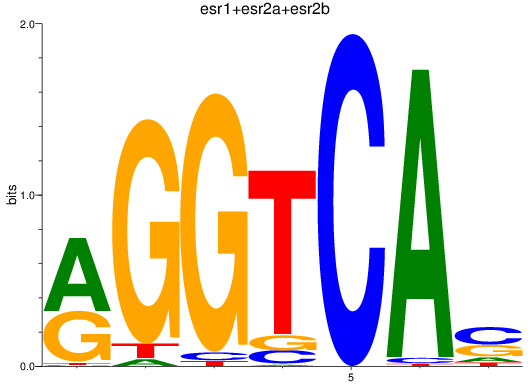
Results for esr1+esr2a+esr2b
Z-value: 1.08
Transcription factors associated with esr1+esr2a+esr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esr1
|
ENSDARG00000004111 | estrogen receptor 1 |
|
esr2a
|
ENSDARG00000016454 | estrogen receptor 2a |
|
esr2b
|
ENSDARG00000034181 | estrogen receptor 2b |
|
esr1
|
ENSDARG00000112357 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| esr2b | dr11_v1_chr13_+_37022601_37022632 | -0.41 | 4.7e-05 | Click! |
| esr2a | dr11_v1_chr20_+_21595244_21595244 | -0.25 | 1.7e-02 | Click! |
| esr1 | dr11_v1_chr20_-_26382284_26382284 | -0.15 | 1.5e-01 | Click! |
Activity profile of esr1+esr2a+esr2b motif
Sorted Z-values of esr1+esr2a+esr2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_55032439 | 8.16 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr5_-_71722257 | 7.71 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr20_+_54738210 | 7.50 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr1_-_59176949 | 6.67 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr25_-_19374710 | 6.08 |
ENSDART00000184483
ENSDART00000188706 |
map1ab
|
microtubule-associated protein 1Ab |
| chr9_+_54644626 | 5.80 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr6_-_40722200 | 5.71 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr5_-_55395964 | 5.57 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr12_-_4683325 | 5.23 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr20_+_5564042 | 5.05 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr12_+_7491690 | 4.99 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr6_+_48618512 | 4.87 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr23_-_45705525 | 4.84 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr16_+_29650698 | 4.84 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr13_+_22480496 | 4.71 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr11_+_7158723 | 4.70 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr17_-_6738538 | 4.67 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr23_+_43954809 | 4.61 |
ENSDART00000164080
|
corin
|
corin, serine peptidase |
| chr24_+_2495197 | 4.59 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr13_-_31435137 | 4.55 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr6_-_11768198 | 4.37 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr2_-_24289641 | 4.36 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr2_-_30460293 | 4.27 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr3_+_39566999 | 4.19 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr23_+_44741500 | 4.18 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr19_-_32783373 | 4.14 |
ENSDART00000145790
|
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr18_-_226800 | 4.12 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr24_-_38374744 | 4.10 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr24_+_25467465 | 4.06 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr5_-_31901468 | 4.01 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr15_+_19544052 | 3.87 |
ENSDART00000062560
|
zgc:77784
|
zgc:77784 |
| chr16_+_52771199 | 3.75 |
ENSDART00000111383
|
baalca
|
brain and acute leukemia, cytoplasmic a |
| chr6_-_9792004 | 3.73 |
ENSDART00000081129
|
cdk15
|
cyclin-dependent kinase 15 |
| chr22_-_13857729 | 3.72 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr25_-_19420949 | 3.71 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr8_+_48613040 | 3.69 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr11_+_11201096 | 3.69 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr4_+_38344 | 3.68 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr9_-_7539297 | 3.68 |
ENSDART00000081550
ENSDART00000081553 |
desma
|
desmin a |
| chr20_+_40150612 | 3.66 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr17_-_12385308 | 3.64 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr13_+_22480857 | 3.62 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr5_+_23624684 | 3.59 |
ENSDART00000051539
|
cx27.5
|
connexin 27.5 |
| chr5_+_64319590 | 3.58 |
ENSDART00000192652
|
FQ377918.1
|
|
| chr16_-_560574 | 3.56 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr3_-_6767440 | 3.54 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr7_+_40228422 | 3.53 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr6_+_36942966 | 3.52 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr1_-_38813679 | 3.52 |
ENSDART00000148917
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr9_+_4306122 | 3.52 |
ENSDART00000193722
ENSDART00000190521 |
kalrna
|
kalirin RhoGEF kinase a |
| chr10_+_22782522 | 3.50 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr5_+_70155935 | 3.50 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr21_-_131236 | 3.49 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr2_-_31302615 | 3.49 |
ENSDART00000034784
ENSDART00000060812 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr12_+_35119762 | 3.43 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr2_+_34767171 | 3.41 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr3_-_1317290 | 3.39 |
ENSDART00000047094
|
LO018552.1
|
|
| chr10_-_17103651 | 3.36 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr5_+_1278092 | 3.28 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr1_-_12278522 | 3.25 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr11_+_5842632 | 3.24 |
ENSDART00000111374
ENSDART00000158599 |
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr2_+_5446087 | 3.23 |
ENSDART00000155165
|
dusp28
|
dual specificity phosphatase 28 |
| chr5_-_23280098 | 3.22 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr19_+_712127 | 3.22 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr5_-_46980651 | 3.20 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr2_-_38000276 | 3.13 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr3_-_13147310 | 3.12 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr25_-_29134654 | 3.09 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr1_-_55810730 | 3.09 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr4_-_191736 | 3.07 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr6_+_10450000 | 3.06 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr23_+_6232895 | 3.05 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr8_+_23213320 | 3.04 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr9_-_31747106 | 3.00 |
ENSDART00000048469
ENSDART00000145204 ENSDART00000186889 |
nalcn
|
sodium leak channel, non-selective |
| chr18_-_14937211 | 2.98 |
ENSDART00000141893
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr12_-_26415499 | 2.98 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr6_-_32025225 | 2.97 |
ENSDART00000006417
|
pgm1
|
phosphoglucomutase 1 |
| chr7_-_51793333 | 2.95 |
ENSDART00000180654
|
BX957362.5
|
|
| chr19_-_9712530 | 2.94 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr24_+_2470061 | 2.94 |
ENSDART00000140383
ENSDART00000191261 |
F13A1 (1 of many)
|
coagulation factor XIII A chain |
| chr6_-_40722480 | 2.93 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr12_+_18556929 | 2.92 |
ENSDART00000191277
|
CT009714.1
|
|
| chr1_+_17676745 | 2.92 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr4_+_17280868 | 2.91 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr16_-_55028740 | 2.88 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr15_-_12500938 | 2.85 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr18_-_16179129 | 2.83 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr9_-_22821901 | 2.82 |
ENSDART00000101711
|
neb
|
nebulin |
| chr23_-_11870962 | 2.82 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr8_+_31821396 | 2.81 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr18_+_16330025 | 2.80 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr6_+_28124393 | 2.80 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr25_+_16945348 | 2.79 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr6_+_3828560 | 2.78 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr14_-_7128980 | 2.76 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr13_-_31452516 | 2.76 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr8_-_49431939 | 2.75 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr15_+_5923851 | 2.74 |
ENSDART00000152520
ENSDART00000145827 ENSDART00000121529 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr9_+_4429593 | 2.72 |
ENSDART00000184855
|
FP015810.1
|
|
| chr7_-_51476276 | 2.72 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr3_+_27770110 | 2.69 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr9_-_22834860 | 2.68 |
ENSDART00000146486
|
neb
|
nebulin |
| chr19_+_30990815 | 2.67 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr9_-_710896 | 2.67 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr7_+_26132665 | 2.67 |
ENSDART00000129834
|
nat16
|
N-acetyltransferase 16 |
| chr3_+_29980603 | 2.64 |
ENSDART00000151509
|
kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr8_+_24861264 | 2.64 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr8_+_14792830 | 2.63 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr10_+_22775253 | 2.63 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr14_-_9277152 | 2.62 |
ENSDART00000189048
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr4_+_4849789 | 2.62 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr20_-_39273987 | 2.61 |
ENSDART00000127173
|
clu
|
clusterin |
| chr23_-_21471022 | 2.60 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr8_+_49778486 | 2.59 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr17_-_44968177 | 2.58 |
ENSDART00000075510
|
ngb
|
neuroglobin |
| chr9_-_49493305 | 2.58 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr6_-_31348999 | 2.58 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr20_+_30490682 | 2.58 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr19_+_30662529 | 2.55 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr3_+_40170216 | 2.53 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr7_-_30174882 | 2.53 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr2_-_10062575 | 2.52 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr21_+_15704556 | 2.52 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr2_-_44720551 | 2.52 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr16_+_5774977 | 2.51 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr10_+_37145007 | 2.51 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr6_+_41463786 | 2.51 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr11_+_42587900 | 2.50 |
ENSDART00000167529
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr20_+_3108597 | 2.50 |
ENSDART00000133435
|
CEP170B (1 of many)
|
si:ch73-212j7.1 |
| chr9_+_42095220 | 2.48 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr18_-_16123222 | 2.48 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr5_-_68916455 | 2.48 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr1_-_25911292 | 2.44 |
ENSDART00000145012
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr6_+_12853655 | 2.44 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr10_+_10636237 | 2.43 |
ENSDART00000136853
|
fam163b
|
family with sequence similarity 163, member B |
| chr18_+_23249519 | 2.42 |
ENSDART00000005740
ENSDART00000147446 ENSDART00000124818 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr3_+_33367954 | 2.41 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr1_-_14332283 | 2.41 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr19_+_41080240 | 2.41 |
ENSDART00000087295
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr1_-_22757145 | 2.39 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr12_-_16084835 | 2.37 |
ENSDART00000090881
|
kcnj19b
|
potassium voltage-gated channel, subfamily J, member 19b |
| chr20_-_25709247 | 2.36 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr25_+_13406069 | 2.36 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr18_-_42313798 | 2.36 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr3_+_39568290 | 2.35 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr23_-_21515182 | 2.34 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr15_-_47838391 | 2.34 |
ENSDART00000180337
|
kbtbd3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr6_-_42112191 | 2.33 |
ENSDART00000085472
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr17_-_40397752 | 2.33 |
ENSDART00000178483
|
BX548062.1
|
|
| chr4_+_5796761 | 2.32 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr11_-_6970107 | 2.32 |
ENSDART00000171255
|
COMP
|
si:ch211-43f4.1 |
| chr17_+_29276544 | 2.32 |
ENSDART00000049399
|
ankrd9
|
ankyrin repeat domain 9 |
| chr11_+_30729745 | 2.30 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr3_+_59117136 | 2.29 |
ENSDART00000182745
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr10_-_31782616 | 2.29 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_+_12236339 | 2.29 |
ENSDART00000132468
|
tpi1b
|
triosephosphate isomerase 1b |
| chr24_-_23974559 | 2.29 |
ENSDART00000080510
ENSDART00000135242 |
ndufb4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4 |
| chr6_-_60031693 | 2.28 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr10_-_39153959 | 2.27 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr1_-_22726233 | 2.23 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr23_-_35082494 | 2.23 |
ENSDART00000189809
|
BX294434.1
|
|
| chr4_+_15981096 | 2.23 |
ENSDART00000122493
|
ucn3l
|
urocortin 3, like |
| chr18_-_15373620 | 2.23 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr4_+_10366532 | 2.21 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr6_+_42587637 | 2.20 |
ENSDART00000179964
|
camkva
|
CaM kinase-like vesicle-associated a |
| chr8_+_14158021 | 2.20 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr4_-_72080351 | 2.20 |
ENSDART00000174925
|
LO017820.1
|
|
| chr14_-_31465905 | 2.20 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr3_-_36260102 | 2.18 |
ENSDART00000126588
|
rac3a
|
Rac family small GTPase 3a |
| chr22_+_18816662 | 2.16 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr3_-_18710009 | 2.16 |
ENSDART00000142478
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr6_-_957830 | 2.16 |
ENSDART00000090019
ENSDART00000184286 |
zeb2b
|
zinc finger E-box binding homeobox 2b |
| chr5_-_29643381 | 2.15 |
ENSDART00000034849
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr5_+_29784172 | 2.15 |
ENSDART00000139035
|
si:ch211-215c18.3
|
si:ch211-215c18.3 |
| chr24_+_39186940 | 2.15 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr12_+_49125510 | 2.14 |
ENSDART00000185804
|
FO704607.1
|
|
| chr20_-_28638871 | 2.14 |
ENSDART00000184779
|
rgs6
|
regulator of G protein signaling 6 |
| chr24_+_29449690 | 2.13 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr3_-_8510201 | 2.13 |
ENSDART00000009151
|
CABZ01064671.1
|
|
| chr13_+_2523032 | 2.12 |
ENSDART00000172261
|
lhb
|
luteinizing hormone, beta polypeptide |
| chr17_-_38778826 | 2.12 |
ENSDART00000168182
ENSDART00000124041 ENSDART00000136921 |
dglucy
|
D-glutamate cyclase |
| chr3_+_34821327 | 2.12 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr1_-_30039331 | 2.10 |
ENSDART00000086935
ENSDART00000143800 |
MARCH4 (1 of many)
|
zgc:153256 |
| chr12_-_10220036 | 2.09 |
ENSDART00000134619
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr18_+_48608366 | 2.08 |
ENSDART00000151229
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr7_-_69983948 | 2.07 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr2_-_42827336 | 2.07 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr21_-_14773692 | 2.06 |
ENSDART00000142145
|
glulc
|
glutamate-ammonia ligase (glutamine synthase) c |
| chr5_+_65491390 | 2.06 |
ENSDART00000159921
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr7_-_22132265 | 2.05 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr2_-_44038698 | 2.05 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr15_+_24691088 | 2.04 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr6_+_49412754 | 2.03 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr4_-_49607096 | 2.03 |
ENSDART00000154381
|
si:dkey-159n16.4
|
si:dkey-159n16.4 |
| chr11_+_30162407 | 2.02 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr17_+_9040165 | 2.02 |
ENSDART00000181221
ENSDART00000181846 |
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_44027391 | 2.02 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr7_-_72427616 | 2.02 |
ENSDART00000181878
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr19_-_30565122 | 2.01 |
ENSDART00000185650
|
hpcal4
|
hippocalcin like 4 |
| chr4_-_20243021 | 2.01 |
ENSDART00000048023
|
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of esr1+esr2a+esr2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 1.2 | 3.7 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 1.1 | 7.7 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 1.0 | 3.1 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.9 | 2.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.9 | 3.7 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.9 | 5.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.9 | 3.5 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.9 | 2.6 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.9 | 2.6 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.8 | 4.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.8 | 3.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.8 | 2.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.7 | 2.9 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.7 | 4.9 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.7 | 3.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.7 | 2.6 | GO:0060405 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.6 | 7.0 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.6 | 1.7 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.6 | 2.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 1.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.5 | 4.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 1.6 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.5 | 1.5 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.5 | 1.5 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.5 | 1.5 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.4 | 0.8 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.4 | 3.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 2.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.4 | 1.6 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 4.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 3.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.4 | 9.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.4 | 0.4 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.4 | 3.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.4 | 8.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 4.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 3.9 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.3 | 2.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.3 | 4.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.3 | 1.7 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.3 | 6.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 4.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 0.9 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.3 | 2.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.3 | 5.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.3 | 2.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 2.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.3 | 1.6 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.3 | 0.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.3 | 4.7 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.3 | 3.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 1.5 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 1.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 6.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 2.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.4 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.2 | 3.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 4.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.7 | GO:2000650 | negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 1.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.9 | GO:1903578 | regulation of oxidative phosphorylation(GO:0002082) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.2 | 1.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 4.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 4.3 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 2.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.2 | 1.8 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.2 | 1.6 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 4.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 0.6 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.2 | 3.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 1.9 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 1.7 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 3.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 2.7 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.2 | 4.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 1.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 0.7 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 3.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 4.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 0.9 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.2 | 0.5 | GO:0097240 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 5.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 0.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 3.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 2.0 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 0.5 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.7 | GO:0072314 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.2 | 1.3 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 3.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 0.8 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.4 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 4.4 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 0.8 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 1.1 | GO:0043589 | skin morphogenesis(GO:0043589) mesenchymal cell migration(GO:0090497) |
| 0.2 | 1.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.9 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 2.4 | GO:0010766 | positive regulation of sodium ion transport(GO:0010765) negative regulation of sodium ion transport(GO:0010766) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.7 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 2.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.4 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.1 | 1.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 1.7 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.8 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.4 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.1 | 0.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 6.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 2.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.7 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.4 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.4 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.1 | 0.5 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 0.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 3.1 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 2.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.5 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 1.2 | GO:0007418 | ventral midline development(GO:0007418) floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 4.8 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 2.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 3.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.6 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 0.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.8 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 1.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 4.2 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.0 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.5 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 4.2 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.8 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 1.5 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.1 | 1.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.6 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.6 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 0.9 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.9 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 11.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 2.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 1.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.8 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.8 | GO:0008343 | adult feeding behavior(GO:0008343) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.3 | GO:0072679 | negative T cell selection(GO:0043383) thymocyte migration(GO:0072679) |
| 0.1 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 2.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 5.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 4.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.6 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 0.6 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 1.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.7 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.1 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.8 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 1.0 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 9.8 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.1 | 1.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.8 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 2.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 2.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.5 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.0 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.1 | 0.7 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 1.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.3 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 3.4 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 1.0 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.1 | 1.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.3 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 9.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 0.8 | GO:0014074 | response to purine-containing compound(GO:0014074) response to ATP(GO:0033198) response to organophosphorus(GO:0046683) |
| 0.1 | 0.3 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.4 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 1.3 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 1.8 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.1 | 1.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.2 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 5.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 3.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 2.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.4 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 1.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 1.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 | 0.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 0.2 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.1 | 1.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.4 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 7.0 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.1 | 0.7 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 1.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 1.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 2.0 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 2.7 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 10.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 3.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 3.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.3 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 2.5 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 2.7 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.9 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 2.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.5 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.1 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 1.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.0 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.6 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.8 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.2 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.3 | GO:0018200 | peptidyl-glutamic acid modification(GO:0018200) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 6.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 1.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 3.0 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.0 | 0.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.7 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 1.1 | GO:0007601 | visual perception(GO:0007601) sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.8 | 4.1 | GO:0043034 | costamere(GO:0043034) |
| 0.8 | 6.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.8 | 3.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.7 | 2.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.6 | 3.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.6 | 3.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.5 | 2.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.5 | 1.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.4 | 2.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 3.0 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 4.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.4 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 7.2 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 3.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.4 | 9.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 3.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 6.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 2.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 2.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 1.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.3 | 1.3 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.3 | 3.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 10.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 1.2 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 3.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 21.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 3.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 11.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 1.0 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 11.2 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.2 | 2.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 0.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 3.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 11.8 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 3.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.4 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 5.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 2.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 5.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.8 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 11.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 2.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 4.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 6.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 1.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 5.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 10.6 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 6.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 8.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.8 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.9 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 1.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 36.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) postsynaptic endosome(GO:0098845) |
| 0.0 | 2.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 9.3 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.8 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.9 | 7.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 3.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.9 | 2.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.9 | 3.5 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.9 | 3.5 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.9 | 2.6 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.7 | 2.9 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.7 | 4.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.7 | 6.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 7.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.6 | 3.6 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.6 | 7.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 2.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.6 | 4.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.6 | 2.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.5 | 2.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 1.5 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.5 | 1.4 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.5 | 3.8 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.4 | 3.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.4 | 8.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.4 | 1.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.4 | 2.9 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 1.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.4 | 1.9 | GO:2001070 | starch binding(GO:2001070) |
| 0.4 | 1.9 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.4 | 3.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.4 | 2.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.4 | 2.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 1.4 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.3 | 7.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 1.7 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.3 | 1.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 2.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 1.5 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.3 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.3 | 2.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 1.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.3 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 1.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 3.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.3 | 0.8 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.3 | 5.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 5.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.3 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 9.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 9.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 0.7 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 4.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 4.1 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 4.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 3.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.3 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 1.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 4.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 2.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 5.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 2.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 1.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 2.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 2.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 0.5 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 4.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 3.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.5 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 2.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 12.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.6 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.2 | 1.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 1.4 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 1.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 8.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.9 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 2.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.7 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.9 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 1.7 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 2.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.4 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 4.5 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 2.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.9 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 1.7 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 6.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.8 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 1.8 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 2.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.5 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 3.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.7 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 1.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 3.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 10.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 5.7 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.1 | 5.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 1.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 2.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 3.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 3.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.3 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 1.0 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 5.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.4 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 0.3 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 44.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.7 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.5 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 2.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 3.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 1.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 8.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 2.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 6.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.3 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 1.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 3.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.9 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0045182 | translation regulator activity(GO:0045182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 3.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 2.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.4 | 5.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 3.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 4.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 2.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 8.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 5.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 16.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 1.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 2.6 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.3 | 7.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 8.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.0 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 1.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 1.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 1.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |