Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
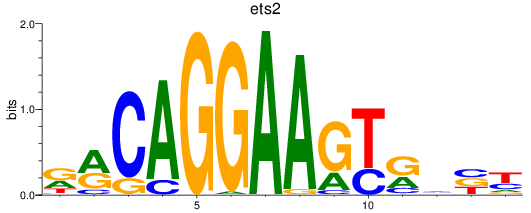
Results for ets2
Z-value: 0.98
Transcription factors associated with ets2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ets2
|
ENSDARG00000103980 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ets2 | dr11_v1_chr10_+_187760_187764 | 0.37 | 2.4e-04 | Click! |
Activity profile of ets2 motif
Sorted Z-values of ets2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25765734 | 10.83 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr15_-_29598679 | 10.53 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr1_-_37383539 | 9.76 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr8_+_3379815 | 9.74 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr15_-_2652640 | 7.12 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr16_+_38201840 | 6.97 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr11_+_25278772 | 6.46 |
ENSDART00000188630
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr1_-_37383741 | 6.37 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr5_-_55981288 | 6.24 |
ENSDART00000146616
|
si:dkey-189h5.6
|
si:dkey-189h5.6 |
| chr15_-_29598444 | 6.13 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr8_-_31107537 | 6.09 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr6_+_112579 | 5.96 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr15_+_15779184 | 5.94 |
ENSDART00000156902
|
si:ch211-33e4.2
|
si:ch211-33e4.2 |
| chr6_-_40657653 | 5.86 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr21_+_30351256 | 5.66 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr16_+_35401543 | 5.66 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr14_-_14643190 | 5.65 |
ENSDART00000167119
|
znf185
|
zinc finger protein 185 with LIM domain |
| chr3_-_27647845 | 5.58 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr5_+_56026031 | 5.48 |
ENSDART00000050970
|
fzd9a
|
frizzled class receptor 9a |
| chr18_+_36631923 | 5.38 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr22_-_1079773 | 5.24 |
ENSDART00000136668
|
si:ch1073-15f12.3
|
si:ch1073-15f12.3 |
| chr17_-_2039511 | 5.21 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr12_+_22576404 | 5.11 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr16_-_25233515 | 5.03 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr21_+_30794351 | 4.94 |
ENSDART00000139486
|
zgc:158225
|
zgc:158225 |
| chr6_-_49063085 | 4.91 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr14_+_20911310 | 4.79 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr7_-_34262080 | 4.75 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr9_-_23807032 | 4.64 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr13_+_30696286 | 4.44 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr4_-_992063 | 4.26 |
ENSDART00000181630
ENSDART00000183898 ENSDART00000160902 |
naga
|
N-acetylgalactosaminidase, alpha |
| chr10_+_8875195 | 4.24 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr1_-_8653385 | 4.17 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr1_-_44796531 | 4.13 |
ENSDART00000144511
|
si:dkey-9i23.6
|
si:dkey-9i23.6 |
| chr10_+_3127809 | 4.06 |
ENSDART00000185500
|
ckmt2a
|
creatine kinase, mitochondrial 2a (sarcomeric) |
| chr10_-_8046764 | 4.03 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr7_-_57637779 | 4.00 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr22_+_19220459 | 3.99 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr4_+_842010 | 3.96 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr21_+_28747069 | 3.86 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr7_-_53117131 | 3.83 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr15_-_3277635 | 3.77 |
ENSDART00000189094
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr3_+_59880317 | 3.74 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr9_-_105135 | 3.68 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr15_+_20239141 | 3.66 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr8_+_44783424 | 3.63 |
ENSDART00000025875
|
si:ch1073-459j12.1
|
si:ch1073-459j12.1 |
| chr17_-_24890843 | 3.60 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr12_-_49149800 | 3.59 |
ENSDART00000182290
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr4_+_12966640 | 3.58 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr17_-_21368775 | 3.58 |
ENSDART00000058027
|
shtn1
|
shootin 1 |
| chr11_+_44135351 | 3.56 |
ENSDART00000182914
|
FO704721.1
|
|
| chr19_-_48312109 | 3.55 |
ENSDART00000161103
|
si:ch73-359m17.9
|
si:ch73-359m17.9 |
| chr1_-_48933 | 3.54 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr4_+_7698862 | 3.53 |
ENSDART00000009909
|
elk3
|
ELK3, ETS-domain protein |
| chr5_+_6672870 | 3.50 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr20_-_35554451 | 3.45 |
ENSDART00000189085
|
adgrf7
|
adhesion G protein-coupled receptor F7 |
| chr9_+_13999620 | 3.41 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr21_+_27448856 | 3.40 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr25_-_13553055 | 3.31 |
ENSDART00000103831
ENSDART00000139290 |
ano10b
|
anoctamin 10b |
| chr13_-_25767210 | 3.29 |
ENSDART00000131792
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr21_+_28747236 | 3.27 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr15_-_43284021 | 3.24 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr10_+_26667475 | 3.23 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr10_+_2582254 | 3.23 |
ENSDART00000016103
|
nxnl2
|
nucleoredoxin like 2 |
| chr19_+_3849378 | 3.18 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr13_-_36582341 | 3.15 |
ENSDART00000137335
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr13_-_33321058 | 3.15 |
ENSDART00000112084
|
si:dkey-71p21.13
|
si:dkey-71p21.13 |
| chr19_-_1855368 | 3.14 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr7_+_73295890 | 3.13 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr9_-_33062891 | 3.12 |
ENSDART00000161182
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr16_-_39570832 | 3.11 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr14_-_9128919 | 3.10 |
ENSDART00000108641
|
sh2d1ab
|
SH2 domain containing 1A duplicate b |
| chr19_+_43297546 | 3.09 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr4_+_76671012 | 3.06 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr18_+_25653599 | 3.04 |
ENSDART00000007856
|
fkbp16
|
FK506 binding protein 16 |
| chr21_+_261490 | 3.03 |
ENSDART00000177919
|
jak2a
|
Janus kinase 2a |
| chr22_-_193234 | 3.02 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr14_-_40797117 | 2.99 |
ENSDART00000122369
|
elf1
|
E74-like ETS transcription factor 1 |
| chr18_+_3140682 | 2.98 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr3_-_16537441 | 2.97 |
ENSDART00000080749
ENSDART00000133824 |
eps8l1
|
eps8-like1 |
| chr9_-_33081781 | 2.96 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr13_-_34858500 | 2.90 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr4_+_76787274 | 2.90 |
ENSDART00000156344
|
si:ch73-56d11.3
|
si:ch73-56d11.3 |
| chr2_+_48282590 | 2.86 |
ENSDART00000035338
|
lpar5a
|
lysophosphatidic acid receptor 5a |
| chr11_+_22109887 | 2.86 |
ENSDART00000122136
|
MDFI
|
si:dkey-91m3.1 |
| chr5_-_42272517 | 2.85 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr10_-_8053385 | 2.80 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr4_+_7677318 | 2.80 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr22_+_20710434 | 2.79 |
ENSDART00000135521
|
peak3
|
PEAK family member 3 |
| chr15_+_12436220 | 2.75 |
ENSDART00000169894
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr23_+_27675581 | 2.75 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr9_-_33081978 | 2.72 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr21_-_25295087 | 2.72 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr8_-_50135492 | 2.69 |
ENSDART00000149398
|
si:ch73-132k15.2
|
si:ch73-132k15.2 |
| chr25_+_7598778 | 2.66 |
ENSDART00000104701
|
FO704779.1
|
|
| chr20_+_54304800 | 2.62 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr6_-_49215790 | 2.62 |
ENSDART00000103376
ENSDART00000132347 ENSDART00000132131 ENSDART00000143252 ENSDART00000065033 |
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr7_+_67467702 | 2.62 |
ENSDART00000168460
ENSDART00000170322 |
rpl13
|
ribosomal protein L13 |
| chr9_-_42484444 | 2.61 |
ENSDART00000048320
ENSDART00000047653 |
tfpia
|
tissue factor pathway inhibitor a |
| chr3_-_36364903 | 2.58 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr17_+_45395846 | 2.57 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr7_-_51639699 | 2.55 |
ENSDART00000128917
|
rps4x
|
ribosomal protein S4, X-linked |
| chr1_+_36674584 | 2.55 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr8_+_25629941 | 2.55 |
ENSDART00000186925
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr8_-_4586696 | 2.54 |
ENSDART00000143511
ENSDART00000134324 |
gp1bb
|
glycoprotein Ib (platelet), beta polypeptide |
| chr5_-_32336613 | 2.54 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_+_59314675 | 2.54 |
ENSDART00000161872
ENSDART00000160658 ENSDART00000169792 ENSDART00000160735 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr12_-_44122412 | 2.53 |
ENSDART00000169094
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr22_+_5687615 | 2.51 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr10_+_9553935 | 2.47 |
ENSDART00000028855
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr20_+_19175031 | 2.47 |
ENSDART00000138028
|
blk
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr3_-_3723494 | 2.47 |
ENSDART00000056023
|
si:dkey-217f16.1
|
si:dkey-217f16.1 |
| chr7_-_66133786 | 2.44 |
ENSDART00000154961
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr11_+_18037729 | 2.43 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr9_-_56232296 | 2.43 |
ENSDART00000149554
|
rpl31
|
ribosomal protein L31 |
| chr7_+_26545502 | 2.40 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr3_+_43826018 | 2.40 |
ENSDART00000166197
ENSDART00000165145 |
litaf
|
lipopolysaccharide-induced TNF factor |
| chr1_-_40735381 | 2.39 |
ENSDART00000093269
|
zgc:153642
|
zgc:153642 |
| chr5_+_50953240 | 2.39 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr16_+_38277339 | 2.38 |
ENSDART00000085143
|
bnipl
|
BCL2 interacting protein like |
| chr1_-_18772241 | 2.38 |
ENSDART00000147645
ENSDART00000133194 ENSDART00000147525 ENSDART00000191493 |
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr14_-_5678457 | 2.37 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr6_+_49881864 | 2.33 |
ENSDART00000075040
|
tubb1
|
tubulin, beta 1 class VI |
| chr5_-_54712159 | 2.33 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr1_-_18848955 | 2.31 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr20_+_54079341 | 2.30 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr18_+_27598755 | 2.30 |
ENSDART00000193808
|
cd82b
|
CD82 molecule b |
| chr5_-_37047147 | 2.30 |
ENSDART00000097712
ENSDART00000158974 |
ftr99
|
finTRIM family, member 99 |
| chr12_-_49168398 | 2.29 |
ENSDART00000186608
|
FO704624.1
|
|
| chr10_-_8053753 | 2.29 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr3_-_34528306 | 2.28 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr11_+_18157260 | 2.28 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr19_+_43523690 | 2.26 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr13_+_15933168 | 2.25 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr11_-_18384534 | 2.20 |
ENSDART00000156499
|
prkcdb
|
protein kinase C, delta b |
| chr13_+_28580357 | 2.18 |
ENSDART00000007211
|
wbp1la
|
WW domain binding protein 1-like a |
| chr23_-_45022681 | 2.18 |
ENSDART00000102640
|
ctc1
|
CTS telomere maintenance complex component 1 |
| chr6_-_1223551 | 2.17 |
ENSDART00000182112
|
CABZ01063034.1
|
|
| chr20_+_54312970 | 2.16 |
ENSDART00000024598
ENSDART00000193172 |
zp2.5
|
zona pellucida glycoprotein 2, tandem duplicate 5 |
| chr20_+_54290356 | 2.15 |
ENSDART00000173347
|
zp2.2
|
zona pellucida glycoprotein 2, tandem duplicate 2 |
| chr5_+_46277593 | 2.14 |
ENSDART00000045598
|
zgc:110626
|
zgc:110626 |
| chr14_+_34501245 | 2.14 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr16_-_10316359 | 2.13 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr11_+_18183220 | 2.13 |
ENSDART00000113468
|
LO018315.10
|
|
| chr13_-_280827 | 2.11 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr19_+_1873059 | 2.10 |
ENSDART00000145246
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr25_+_5068442 | 2.10 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr15_+_436776 | 2.07 |
ENSDART00000008504
|
med17
|
mediator complex subunit 17 |
| chr19_+_48024457 | 2.07 |
ENSDART00000163823
|
kpnb1
|
karyopherin (importin) beta 1 |
| chr13_-_22961605 | 2.05 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr9_-_9419704 | 2.05 |
ENSDART00000138996
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr14_-_897874 | 2.04 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr3_+_56574623 | 2.02 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr1_+_26622214 | 2.01 |
ENSDART00000182040
ENSDART00000152643 |
coro2a
|
coronin, actin binding protein, 2A |
| chr20_+_54309148 | 2.00 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr7_-_24112484 | 2.00 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr3_-_45777226 | 1.98 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr20_-_26937453 | 1.97 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr11_+_18053333 | 1.96 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr1_+_38142715 | 1.95 |
ENSDART00000079928
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr4_-_149334 | 1.95 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr19_+_47476908 | 1.93 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr11_+_141504 | 1.93 |
ENSDART00000086166
|
NCKAP1L
|
zgc:172352 |
| chr21_-_3796461 | 1.92 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr5_+_40837539 | 1.92 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr20_+_54299419 | 1.92 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr14_-_52408307 | 1.90 |
ENSDART00000167045
ENSDART00000162719 ENSDART00000160293 |
polr2b
|
polymerase (RNA) II (DNA directed) polypeptide B |
| chr17_+_132555 | 1.89 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr22_-_22337382 | 1.88 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr3_-_16719244 | 1.88 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr10_-_20650302 | 1.88 |
ENSDART00000142028
ENSDART00000064662 |
rassf2b
|
Ras association (RalGDS/AF-6) domain family member 2b |
| chr14_+_23709543 | 1.87 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr9_+_31752628 | 1.87 |
ENSDART00000060054
|
itgbl1
|
integrin, beta-like 1 |
| chr3_+_23742868 | 1.87 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr4_+_77957611 | 1.85 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr16_-_40426837 | 1.85 |
ENSDART00000193690
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr3_-_61116258 | 1.84 |
ENSDART00000009194
ENSDART00000156978 |
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr7_-_54098631 | 1.83 |
ENSDART00000165499
|
rps17
|
ribosomal protein S17 |
| chr16_+_13965923 | 1.83 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr7_-_26603743 | 1.82 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr9_+_13985567 | 1.82 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr15_+_11883804 | 1.81 |
ENSDART00000163255
|
gpr184
|
G protein-coupled receptor 184 |
| chr25_-_37465064 | 1.81 |
ENSDART00000186128
|
zgc:158366
|
zgc:158366 |
| chr19_-_79202 | 1.80 |
ENSDART00000166009
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr12_+_763051 | 1.80 |
ENSDART00000152579
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr19_-_43757568 | 1.79 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr11_+_18130300 | 1.79 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr23_+_9560797 | 1.79 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr8_+_21254192 | 1.79 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr7_-_66877058 | 1.78 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr2_+_22495274 | 1.78 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 VRAC subunit Da |
| chr9_+_48819280 | 1.77 |
ENSDART00000112555
|
spc25
|
SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr23_-_19682971 | 1.77 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr5_+_21181047 | 1.76 |
ENSDART00000088506
|
arhgap25
|
Rho GTPase activating protein 25 |
| chr16_-_9802449 | 1.76 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr2_+_13069168 | 1.76 |
ENSDART00000192832
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr11_+_8129536 | 1.76 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr6_+_39360377 | 1.75 |
ENSDART00000028260
ENSDART00000151322 |
zgc:77517
|
zgc:77517 |
| chr19_+_3206263 | 1.74 |
ENSDART00000020344
|
zgc:86598
|
zgc:86598 |
| chr8_-_17184482 | 1.72 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ets2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.4 | 5.7 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 1.3 | 5.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 1.3 | 3.8 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.9 | 4.3 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.8 | 2.5 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.8 | 2.5 | GO:0070228 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.8 | 4.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.7 | 3.6 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.7 | 2.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.7 | 2.0 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.6 | 6.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.6 | 2.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.6 | 4.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.6 | 3.6 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.6 | 2.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.5 | 2.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.5 | 1.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.5 | 2.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 1.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.5 | 2.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.4 | 2.5 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.4 | 2.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.4 | 1.2 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.4 | 8.6 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.4 | 1.9 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.4 | 17.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.3 | 2.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.7 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.3 | 1.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 10.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.3 | 2.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 4.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 2.5 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.3 | 2.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 1.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 4.1 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 1.6 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.3 | 0.9 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.3 | 4.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 1.1 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.3 | 2.8 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 1.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.3 | 0.8 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.3 | 0.8 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.3 | 4.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 1.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 2.9 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 1.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 1.2 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.2 | 2.6 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 0.7 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 0.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 2.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 0.8 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.2 | 1.5 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 0.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.2 | 6.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.2 | 1.6 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.2 | 7.0 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.2 | 5.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 1.5 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.2 | 0.7 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.2 | 1.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 2.6 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.2 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 1.5 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 2.2 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.4 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 2.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 1.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 3.1 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 7.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 2.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 5.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.9 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 1.5 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.2 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 3.4 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 4.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.4 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.9 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 3.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.6 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.3 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.1 | 1.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 0.6 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 7.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 6.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 3.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.8 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 0.5 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.7 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 1.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.7 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.0 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.1 | 0.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.5 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.7 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.3 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.8 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.3 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.1 | 1.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 3.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.0 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 5.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.3 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 3.7 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 2.6 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.1 | 4.8 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 1.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.8 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 1.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.7 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 2.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 3.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.8 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.7 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 2.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 2.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 1.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 2.0 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.6 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.1 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 3.3 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 1.0 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.4 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.9 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.4 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 3.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.0 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.4 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 1.5 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 5.6 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.0 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 6.9 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0051597 | release of cytochrome c from mitochondria(GO:0001836) response to methylmercury(GO:0051597) |
| 0.0 | 0.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.9 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 3.5 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.0 | GO:0010469 | regulation of receptor activity(GO:0010469) |
| 0.0 | 0.9 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.9 | GO:0001754 | eye photoreceptor cell differentiation(GO:0001754) |
| 0.0 | 1.6 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.5 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.9 | 3.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.8 | 4.2 | GO:0097433 | dense body(GO:0097433) |
| 0.8 | 2.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.7 | 2.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.7 | 2.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.6 | 2.4 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.6 | 3.6 | GO:0030891 | VCB complex(GO:0030891) |
| 0.6 | 1.7 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.5 | 4.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.5 | 2.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.4 | 2.6 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.4 | 2.0 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.4 | 0.8 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.4 | 1.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.4 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.3 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 1.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 0.9 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 3.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 3.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 2.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.3 | 2.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 2.9 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.3 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 7.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 1.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 1.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 1.7 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 0.6 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 0.8 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 1.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 3.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 1.0 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.2 | 9.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 2.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 0.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 9.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 7.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 16.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.9 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 12.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 16.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.4 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 1.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.5 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 4.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.4 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 0.3 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 1.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.4 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 10.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 5.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.2 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 2.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.8 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 1.0 | GO:0098844 | dendritic spine head(GO:0044327) postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 0.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 7.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 6.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 11.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.8 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 2.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 4.3 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.2 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 2.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.0 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 6.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 30.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 1.2 | 6.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 1.2 | 3.6 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 1.2 | 4.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.9 | 4.3 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.9 | 2.6 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.8 | 4.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.6 | 6.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.6 | 3.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.6 | 1.7 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.5 | 0.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.5 | 1.6 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.5 | 3.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.5 | 7.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.5 | 0.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.5 | 5.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 3.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.4 | 1.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 1.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 2.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.4 | 2.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.4 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 5.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 4.1 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 6.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 0.8 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.3 | 2.9 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.3 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 0.8 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.3 | 1.0 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.3 | 2.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 2.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.6 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.2 | 1.5 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 3.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 2.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 1.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 3.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 2.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 2.6 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 6.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.9 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 1.4 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 8.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 3.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 1.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 1.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 1.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 2.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 6.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 2.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.2 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 2.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 20.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.7 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 8.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.4 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 4.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 2.2 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 9.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.5 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.6 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.8 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.5 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 2.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 6.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 16.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 3.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 8.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 9.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 6.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.8 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 2.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 4.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 7.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 3.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 6.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.5 | 2.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.4 | 2.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 7.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.3 | 2.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 1.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 10.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 4.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 4.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 1.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 1.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 3.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 9.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 2.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 1.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 9.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 4.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.4 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 4.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.3 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 1.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 1.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |