Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
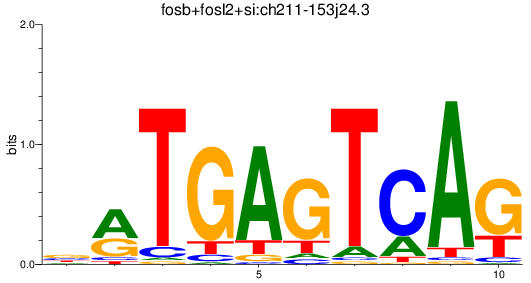
Results for fosb+fosl2+si:ch211-153j24.3
Z-value: 1.16
Transcription factors associated with fosb+fosl2+si:ch211-153j24.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2
|
ENSDARG00000040623 | fos-like antigen 2 |
|
fosb
|
ENSDARG00000055751 | FBJ murine osteosarcoma viral oncogene homolog B |
|
si_ch211-153j24.3
|
ENSDARG00000068428 | si_ch211-153j24.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-153j24.3 | dr11_v1_chr20_+_46560258_46560258 | -0.52 | 5.6e-08 | Click! |
| fosl2 | dr11_v1_chr17_+_41302660_41302660 | 0.13 | 2.0e-01 | Click! |
| fosb | dr11_v1_chr18_+_36770166_36770166 | 0.08 | 4.5e-01 | Click! |
Activity profile of fosb+fosl2+si:ch211-153j24.3 motif
Sorted Z-values of fosb+fosl2+si:ch211-153j24.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_5595546 | 13.91 |
ENSDART00000191825
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr25_-_22187397 | 13.22 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr25_+_13191391 | 12.46 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr21_-_2415808 | 11.70 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr16_+_23403602 | 10.99 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr14_-_11456724 | 10.27 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr1_+_14253118 | 10.17 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr16_-_22930925 | 9.56 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr7_+_25033924 | 9.47 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr11_-_24681292 | 9.35 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr19_+_7567763 | 8.74 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr17_-_53359028 | 8.18 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr20_+_26880668 | 8.07 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr2_+_30379650 | 7.93 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr21_+_5636008 | 7.86 |
ENSDART00000158385
|
shroom3
|
shroom family member 3 |
| chr11_-_18253111 | 7.77 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr25_+_13191615 | 7.67 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr12_-_15205087 | 7.55 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr12_+_25945560 | 7.22 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr14_-_1538600 | 7.18 |
ENSDART00000180925
|
CABZ01109480.1
|
|
| chr3_-_48259289 | 7.18 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr8_+_6576940 | 6.86 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr9_+_426392 | 6.77 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr5_-_29531948 | 6.71 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr23_+_4324625 | 6.60 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr12_-_212843 | 6.34 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr6_-_35439406 | 6.12 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr16_-_21785261 | 5.97 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr4_+_16715267 | 5.95 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr1_+_59073436 | 5.92 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr16_+_23960744 | 5.89 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr5_-_25582721 | 5.88 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr2_+_31671545 | 5.86 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr10_+_4987766 | 5.82 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr3_+_32118670 | 5.77 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr22_-_15593824 | 5.68 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr17_+_21452258 | 5.65 |
ENSDART00000157098
|
pla2g4f.1
|
phospholipase A2, group IVF, tandem duplicate 1 |
| chr6_+_7507390 | 5.65 |
ENSDART00000149285
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr7_+_33172066 | 5.64 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr3_-_32859335 | 5.56 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr19_+_4061699 | 5.55 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr10_+_187760 | 5.49 |
ENSDART00000161007
ENSDART00000160651 |
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr3_+_1179601 | 5.36 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr25_-_22191733 | 5.35 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr2_-_985417 | 5.33 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr24_+_264839 | 5.28 |
ENSDART00000066872
|
ccr12b.2
|
chemokine (C-C motif) receptor 12b, tandem duplicate 2 |
| chr15_+_963292 | 5.28 |
ENSDART00000156586
|
alox5b.2
|
arachidonate 5-lipoxygenase b, tandem duplicate 2 |
| chr9_-_9992697 | 5.16 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr19_+_348729 | 5.12 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr13_-_37122217 | 5.08 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr19_+_2670130 | 4.96 |
ENSDART00000152152
|
si:ch73-24k9.2
|
si:ch73-24k9.2 |
| chr9_-_21838045 | 4.88 |
ENSDART00000147471
|
acod1
|
aconitate decarboxylase 1 |
| chr23_+_1730663 | 4.77 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr4_+_25181572 | 4.77 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr4_-_77216726 | 4.75 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr7_-_32981559 | 4.67 |
ENSDART00000175614
|
pkp3b
|
plakophilin 3b |
| chr16_+_49005321 | 4.66 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr6_-_442163 | 4.63 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr1_+_41588170 | 4.62 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr22_+_16497670 | 4.60 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr7_+_17443567 | 4.55 |
ENSDART00000060383
|
nitr2b
|
novel immune-type receptor 2b |
| chr6_+_39108858 | 4.53 |
ENSDART00000154232
ENSDART00000192092 |
prss60.3
|
protease, serine, 60.3 |
| chr14_-_34605607 | 4.52 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr6_-_41091151 | 4.51 |
ENSDART00000154963
ENSDART00000153818 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr20_+_7084154 | 4.51 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr23_+_19213472 | 4.47 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr15_-_4568154 | 4.34 |
ENSDART00000155254
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr14_+_33882973 | 4.33 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr5_+_35398745 | 4.32 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr7_+_15736230 | 4.27 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr19_-_7420867 | 4.21 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr5_+_38200807 | 4.20 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr22_-_22337382 | 4.18 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr8_+_44358443 | 4.11 |
ENSDART00000189130
ENSDART00000189212 |
CU914776.2
|
|
| chr8_-_20291922 | 4.11 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr7_-_20611039 | 4.10 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr17_+_32623931 | 4.10 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr11_+_37638873 | 4.07 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr2_-_41861040 | 4.03 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr1_-_47122058 | 4.00 |
ENSDART00000159925
ENSDART00000101143 ENSDART00000176803 |
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr13_-_15982707 | 3.95 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr5_+_24089334 | 3.85 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr19_+_37118547 | 3.81 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr8_-_18010735 | 3.79 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr2_+_15100742 | 3.73 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr18_-_45617146 | 3.73 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr7_-_25697285 | 3.72 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr4_+_77933084 | 3.66 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr25_+_4954220 | 3.63 |
ENSDART00000156034
|
si:ch73-265h17.2
|
si:ch73-265h17.2 |
| chr21_+_11778823 | 3.60 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr22_+_19247255 | 3.60 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr23_-_42752387 | 3.51 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr4_-_13518381 | 3.49 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr17_-_26868169 | 3.48 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr23_+_5524247 | 3.46 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr24_-_33366188 | 3.44 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr4_-_41582875 | 3.41 |
ENSDART00000182308
ENSDART00000172173 ENSDART00000163135 |
si:dkeyp-44d3.5
|
si:dkeyp-44d3.5 |
| chr5_-_1047504 | 3.40 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr25_+_32474031 | 3.39 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr8_-_7603516 | 3.33 |
ENSDART00000179826
ENSDART00000190153 |
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr3_-_8765165 | 3.31 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr9_+_20483846 | 3.26 |
ENSDART00000192067
ENSDART00000145111 |
parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr8_+_28695914 | 3.25 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr23_-_30045661 | 3.22 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr5_-_67762434 | 3.18 |
ENSDART00000167301
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr10_+_28428222 | 3.17 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr5_+_54400971 | 3.14 |
ENSDART00000169695
|
bspry
|
B-box and SPRY domain containing |
| chr5_+_42064144 | 3.14 |
ENSDART00000035235
|
si:ch211-202a12.4
|
si:ch211-202a12.4 |
| chr1_-_59139599 | 3.13 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr15_-_37733238 | 3.12 |
ENSDART00000115178
|
si:dkey-42l23.7
|
si:dkey-42l23.7 |
| chr6_+_56147812 | 3.09 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr16_-_24832038 | 3.08 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr25_-_173165 | 3.06 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr25_+_29474583 | 3.06 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr23_-_39636195 | 3.05 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr18_+_50526290 | 3.04 |
ENSDART00000175194
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr14_-_34605804 | 3.00 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr13_+_18533005 | 2.96 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr12_-_22355430 | 2.90 |
ENSDART00000153296
ENSDART00000056919 ENSDART00000159036 |
nsfb
|
N-ethylmaleimide-sensitive factor b |
| chr2_-_3437862 | 2.87 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr13_-_51846224 | 2.87 |
ENSDART00000184663
|
LT631684.2
|
|
| chr24_+_260902 | 2.86 |
ENSDART00000151992
|
ccr12b.1
|
chemokine (C-C motif) receptor 12b, tandem duplicate 1 |
| chr11_-_27821 | 2.84 |
ENSDART00000158769
ENSDART00000172970 ENSDART00000173118 ENSDART00000168674 ENSDART00000163545 ENSDART00000173411 ENSDART00000172132 |
sp1
|
sp1 transcription factor |
| chr23_-_1017605 | 2.84 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr16_-_28878080 | 2.81 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr15_-_38129845 | 2.81 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr3_+_29476085 | 2.77 |
ENSDART00000184495
ENSDART00000181058 |
fam83fa
|
family with sequence similarity 83, member Fa |
| chr8_-_7603700 | 2.76 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr5_+_1933131 | 2.75 |
ENSDART00000061693
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr22_+_19218733 | 2.75 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr24_-_6078222 | 2.74 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr19_+_24374196 | 2.72 |
ENSDART00000140732
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr7_+_14005111 | 2.61 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr5_-_43959972 | 2.60 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr5_-_9760173 | 2.60 |
ENSDART00000172523
|
ddr2l
|
discoidin domain receptor family, member 2, like |
| chr16_-_33816082 | 2.58 |
ENSDART00000181769
|
rspo1
|
R-spondin 1 |
| chr4_+_77971104 | 2.58 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr21_-_28640316 | 2.54 |
ENSDART00000128237
|
nrg2a
|
neuregulin 2a |
| chr25_+_23336310 | 2.53 |
ENSDART00000156457
|
ptprjb.2
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 2 |
| chr24_-_23998897 | 2.52 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr6_-_37744430 | 2.52 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr22_-_10110959 | 2.51 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr23_-_1017428 | 2.51 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr4_-_19742300 | 2.50 |
ENSDART00000066964
ENSDART00000100952 |
hgfa
|
hepatocyte growth factor a |
| chr25_+_13205878 | 2.49 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr11_+_13630107 | 2.46 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr13_+_23988442 | 2.45 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr3_-_21094437 | 2.37 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr8_-_22288258 | 2.34 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr12_+_48784731 | 2.34 |
ENSDART00000158348
|
zmiz1b
|
zinc finger, MIZ-type containing 1b |
| chr5_+_4564233 | 2.32 |
ENSDART00000193435
|
CABZ01058647.1
|
|
| chr17_+_6667441 | 2.32 |
ENSDART00000123503
ENSDART00000180381 ENSDART00000156140 |
smc6
|
structural maintenance of chromosomes 6 |
| chr25_+_8955530 | 2.32 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr4_-_75505048 | 2.29 |
ENSDART00000163665
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr11_+_10975012 | 2.29 |
ENSDART00000192872
|
itgb6
|
integrin, beta 6 |
| chr25_+_13498188 | 2.28 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr8_+_8671229 | 2.27 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr7_+_56453646 | 2.25 |
ENSDART00000112483
|
slc22a31
|
solute carrier family 22, member 31 |
| chr16_-_51288178 | 2.23 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr6_+_51713076 | 2.22 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr12_-_19250854 | 2.21 |
ENSDART00000152844
|
cdc42ep1a
|
CDC42 effector protein (Rho GTPase binding) 1a |
| chr3_+_17616201 | 2.21 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr15_-_41382344 | 2.20 |
ENSDART00000190445
ENSDART00000189776 |
si:dkey-121n8.7
|
si:dkey-121n8.7 |
| chr11_+_21050326 | 2.19 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr8_-_38105053 | 2.18 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr19_-_33212023 | 2.17 |
ENSDART00000189209
|
trib1
|
tribbles pseudokinase 1 |
| chr21_+_45316330 | 2.17 |
ENSDART00000056474
ENSDART00000149314 ENSDART00000149272 ENSDART00000149156 ENSDART00000099497 |
tcf7
|
transcription factor 7 |
| chr4_+_70191443 | 2.17 |
ENSDART00000164404
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr13_-_50567658 | 2.15 |
ENSDART00000132281
|
blnk
|
B cell linker |
| chr15_+_42559910 | 2.14 |
ENSDART00000075785
|
cldn17
|
claudin 17 |
| chr20_-_49889111 | 2.13 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr4_+_68939074 | 2.11 |
ENSDART00000180232
|
si:dkey-264f17.2
|
si:dkey-264f17.2 |
| chr23_-_6865946 | 2.10 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr22_+_39084829 | 2.08 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr16_-_16481046 | 2.08 |
ENSDART00000158704
|
nbeal2
|
neurobeachin-like 2 |
| chr15_-_31177324 | 2.06 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr16_+_25245857 | 2.06 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr4_+_77735212 | 2.05 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr3_-_34069637 | 2.04 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr4_+_77948970 | 2.02 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr11_+_13629528 | 2.02 |
ENSDART00000186509
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr7_-_5125799 | 2.02 |
ENSDART00000173390
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr1_-_33380340 | 2.01 |
ENSDART00000181515
|
cd99
|
CD99 molecule |
| chr3_-_584950 | 2.00 |
ENSDART00000164752
|
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr4_-_32456788 | 1.99 |
ENSDART00000151862
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr14_-_16807206 | 1.97 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr24_+_42074143 | 1.96 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr19_+_42847306 | 1.95 |
ENSDART00000135164
|
pdcd6ip
|
programmed cell death 6 interacting protein |
| chr4_-_75899294 | 1.95 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr1_-_59411901 | 1.93 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr11_-_23501467 | 1.92 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr4_-_70131926 | 1.91 |
ENSDART00000108511
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr2_-_6519017 | 1.91 |
ENSDART00000181716
|
rgs1
|
regulator of G protein signaling 1 |
| chr14_+_21699414 | 1.88 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr17_-_15600455 | 1.85 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr21_-_21020708 | 1.80 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr4_+_5317483 | 1.79 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr1_+_55452892 | 1.78 |
ENSDART00000122508
|
CR788255.1
|
|
| chr19_+_15441022 | 1.77 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr7_+_20298333 | 1.77 |
ENSDART00000023089
ENSDART00000131019 |
acadvl
|
acyl-CoA dehydrogenase very long chain |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosb+fosl2+si:ch211-153j24.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 2.0 | 5.9 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 1.6 | 7.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 1.5 | 4.5 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 1.3 | 3.9 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 1.1 | 3.4 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 1.1 | 11.7 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 1.0 | 10.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 1.0 | 5.9 | GO:0033031 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.0 | 5.8 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.9 | 3.7 | GO:0042117 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.9 | 4.3 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.9 | 6.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.8 | 4.2 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.8 | 4.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.7 | 2.2 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.7 | 2.9 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.7 | 4.9 | GO:1903426 | regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.7 | 6.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.7 | 5.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.6 | 9.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.6 | 5.7 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.5 | 1.6 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.5 | 2.2 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.5 | 2.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.5 | 1.4 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.4 | 5.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 1.7 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 1.9 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.4 | 4.1 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 2.5 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.4 | 8.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 2.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 2.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.3 | 7.9 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.3 | 7.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 1.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 3.1 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 3.5 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.2 | 0.9 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 2.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 1.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 2.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 3.9 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.2 | 0.7 | GO:0009750 | response to fructose(GO:0009750) |
| 0.2 | 2.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 4.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 5.4 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.2 | 5.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 3.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 2.6 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 1.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 4.2 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.2 | 1.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 2.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 1.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 2.9 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.2 | 1.1 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 1.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 15.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 2.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 2.0 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 3.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.9 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 18.5 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 13.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 4.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.4 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 2.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.6 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.1 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 1.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 2.0 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 4.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 2.5 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 3.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 4.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 4.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.9 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 2.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 1.5 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 3.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 2.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 1.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 5.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.7 | GO:0021986 | habenula development(GO:0021986) |
| 0.1 | 0.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 2.6 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 2.7 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 3.6 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 1.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 1.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 2.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 3.1 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 3.8 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 2.2 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.2 | GO:0070935 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 5.1 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 2.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.5 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 5.2 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 3.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 2.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 3.3 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 1.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.9 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 1.2 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 1.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.3 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 4.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.5 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 4.7 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.3 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 2.2 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 4.2 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
| 0.0 | 1.1 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.9 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.0 | GO:0048545 | response to steroid hormone(GO:0048545) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.2 | 23.2 | GO:0030057 | desmosome(GO:0030057) |
| 1.2 | 3.5 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.8 | 2.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.6 | 4.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.5 | 2.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.5 | 6.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.5 | 7.5 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 5.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.4 | 7.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 2.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 3.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 1.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 3.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 7.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 4.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 6.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 2.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 0.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.2 | 1.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 35.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 5.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 18.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 7.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 5.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 4.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 7.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 5.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 6.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 6.3 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 5.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 5.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.8 | GO:0098791 | trans-Golgi network(GO:0005802) Golgi subcompartment(GO:0098791) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 6.1 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 2.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 26.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 2.3 | 13.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 2.0 | 5.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.3 | 6.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 1.1 | 5.6 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 1.1 | 3.4 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 1.0 | 6.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 1.0 | 5.9 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.0 | 2.9 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.8 | 7.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.8 | 8.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.8 | 5.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.7 | 2.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.7 | 19.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.6 | 4.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 1.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.5 | 3.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.5 | 2.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.5 | 2.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.5 | 11.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 4.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 1.7 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.4 | 1.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.4 | 2.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 14.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 1.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 1.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 2.6 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.3 | 2.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 1.4 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 2.5 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 3.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 24.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 4.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 5.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 3.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 10.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.2 | 4.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.2 | 4.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.2 | 3.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 0.7 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 2.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 1.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 0.9 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 3.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.7 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 2.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 2.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 7.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 3.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 8.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 6.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 7.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 5.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 1.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 3.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 5.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 18.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 2.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 2.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.7 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 6.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 14.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 6.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 9.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.8 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 2.0 | GO:0008134 | transcription factor binding(GO:0008134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 3.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 2.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 5.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 7.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.4 | 2.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.4 | 3.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 3.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 2.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 2.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 3.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 5.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 2.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 3.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |