Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
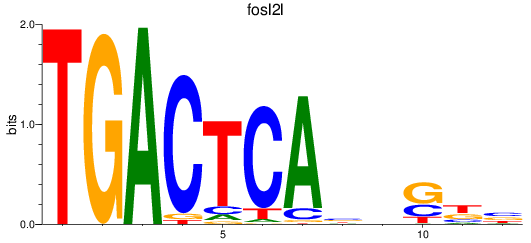
Results for fosl2l
Z-value: 0.52
Transcription factors associated with fosl2l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2l
|
ENSDARG00000092174 | fos-like antigen 2 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-59d15.4 | dr11_v1_chr20_+_9211237_9211237 | -0.11 | 2.9e-01 | Click! |
Activity profile of fosl2l motif
Sorted Z-values of fosl2l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_20381485 | 4.95 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr1_-_56176976 | 3.68 |
ENSDART00000052688
|
c3a.1
|
complement component c3a, duplicate 1 |
| chr20_+_26880668 | 3.39 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr20_-_35578435 | 3.38 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr13_+_23988442 | 3.08 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr8_-_38201415 | 3.07 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr3_+_21189766 | 3.05 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr4_-_9891874 | 2.90 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr13_+_2908764 | 2.87 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr16_+_23403602 | 2.83 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr25_+_10410620 | 2.60 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr7_+_33172066 | 2.24 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr8_+_8671229 | 2.23 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr13_+_4671698 | 2.23 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr15_+_21202820 | 2.20 |
ENSDART00000154036
|
si:dkey-52d15.2
|
si:dkey-52d15.2 |
| chr16_+_49005321 | 2.14 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr6_-_609880 | 2.14 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr6_+_54711306 | 2.11 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr20_+_28364742 | 2.08 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr8_+_6576940 | 2.04 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr7_+_19482084 | 1.97 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr22_+_38194151 | 1.93 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr2_+_31671545 | 1.91 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr23_-_1017605 | 1.88 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr9_+_46745348 | 1.87 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr8_+_22516728 | 1.85 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr23_-_1017428 | 1.72 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr7_-_7398350 | 1.71 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr19_-_7420867 | 1.69 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr1_-_25177086 | 1.69 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr4_+_16715267 | 1.65 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr3_+_4346854 | 1.59 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr8_+_1766206 | 1.58 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr24_+_24726956 | 1.52 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr17_+_6585333 | 1.49 |
ENSDART00000124293
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr4_+_25181572 | 1.49 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr16_+_32136550 | 1.48 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr3_-_48259289 | 1.46 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr5_+_22370167 | 1.46 |
ENSDART00000145163
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr15_+_42235449 | 1.46 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr7_+_39706004 | 1.45 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr3_-_4303262 | 1.43 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr24_-_33366188 | 1.42 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr8_-_18667693 | 1.42 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr25_+_18583877 | 1.41 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr20_-_23439011 | 1.38 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr3_+_46762703 | 1.37 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr3_-_26017592 | 1.34 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr20_-_34127415 | 1.33 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr24_+_38216340 | 1.31 |
ENSDART00000188561
|
CU896602.4
|
|
| chr25_+_32474031 | 1.31 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr16_-_31598771 | 1.30 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr4_-_18775548 | 1.30 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr20_-_34670236 | 1.29 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr16_+_26547152 | 1.28 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr19_+_37120491 | 1.28 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr3_+_32118670 | 1.27 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr7_-_29534001 | 1.25 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr6_-_59357256 | 1.25 |
ENSDART00000074534
|
fam210b
|
family with sequence similarity 210, member B |
| chr23_+_28378543 | 1.25 |
ENSDART00000145327
|
zgc:153867
|
zgc:153867 |
| chr8_-_11170114 | 1.25 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr15_+_3766101 | 1.24 |
ENSDART00000042580
ENSDART00000112698 ENSDART00000187035 ENSDART00000165571 ENSDART00000121752 |
RNF14 (1 of many)
|
zmp:0000000524 |
| chr5_+_51026563 | 1.20 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr19_+_24488403 | 1.19 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr3_+_34121156 | 1.19 |
ENSDART00000174929
|
aldh3b1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr7_-_30926030 | 1.18 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr3_-_26017831 | 1.16 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_39190317 | 1.15 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr25_-_33275666 | 1.14 |
ENSDART00000193605
|
CABZ01046954.1
|
|
| chr2_-_13691834 | 1.14 |
ENSDART00000186570
|
CABZ01044235.1
|
|
| chr2_-_985417 | 1.12 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr19_-_7043355 | 1.10 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr13_-_10945288 | 1.09 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr14_+_33882973 | 1.09 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr24_+_38223618 | 1.08 |
ENSDART00000003495
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr7_+_14005111 | 1.06 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr17_-_15600455 | 1.05 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr4_-_76303301 | 1.05 |
ENSDART00000158504
|
zgc:174944
|
zgc:174944 |
| chr4_-_17822447 | 1.03 |
ENSDART00000185502
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr7_+_69019851 | 1.03 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr25_-_11378623 | 1.02 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr23_-_45398622 | 1.02 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr25_+_8407892 | 1.00 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr4_-_76105527 | 1.00 |
ENSDART00000159108
|
zgc:110171
|
zgc:110171 |
| chr13_-_49585385 | 0.96 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr5_-_1047504 | 0.96 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr15_+_17343319 | 0.95 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr18_+_25225524 | 0.94 |
ENSDART00000055563
|
si:dkeyp-59c12.1
|
si:dkeyp-59c12.1 |
| chr4_+_76973771 | 0.93 |
ENSDART00000142978
|
si:dkey-240n22.3
|
si:dkey-240n22.3 |
| chr4_+_58667348 | 0.92 |
ENSDART00000186596
ENSDART00000180673 |
CR388148.2
|
|
| chr22_+_17399124 | 0.92 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr21_-_27338639 | 0.92 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr19_-_7070691 | 0.91 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr20_+_6543674 | 0.91 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr6_-_40842768 | 0.89 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr15_-_18197008 | 0.86 |
ENSDART00000147632
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr3_+_2754885 | 0.86 |
ENSDART00000188692
|
CR388047.2
|
|
| chr8_-_22288004 | 0.86 |
ENSDART00000100042
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr24_+_38208823 | 0.86 |
ENSDART00000187538
ENSDART00000190189 |
CU896602.3
|
|
| chr16_-_21785261 | 0.85 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr22_-_6884981 | 0.84 |
ENSDART00000124219
|
FO904898.4
|
|
| chr10_-_32877348 | 0.84 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr23_-_4704938 | 0.84 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr10_-_44482911 | 0.84 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr2_+_25378457 | 0.82 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr24_+_38201089 | 0.81 |
ENSDART00000132338
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr4_-_75651505 | 0.81 |
ENSDART00000115143
|
si:dkey-71l4.2
|
si:dkey-71l4.2 |
| chr4_+_75575252 | 0.81 |
ENSDART00000166536
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr4_+_58661331 | 0.80 |
ENSDART00000188763
|
CR388148.2
|
|
| chr21_+_22892836 | 0.80 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr12_-_17147473 | 0.78 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr4_+_68519945 | 0.78 |
ENSDART00000171148
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr13_-_33134611 | 0.78 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr3_+_43955864 | 0.77 |
ENSDART00000168267
|
BX571715.1
|
|
| chr1_+_46598764 | 0.76 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr25_+_13498188 | 0.76 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr13_+_19283936 | 0.76 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr11_+_37905630 | 0.75 |
ENSDART00000170303
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr1_+_10318089 | 0.75 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr11_+_44617021 | 0.75 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr4_-_76186742 | 0.74 |
ENSDART00000163970
|
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr2_+_30379650 | 0.74 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr7_-_20103384 | 0.74 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr8_+_30501170 | 0.73 |
ENSDART00000137009
|
si:dkey-91m11.5
|
si:dkey-91m11.5 |
| chr20_-_21994901 | 0.73 |
ENSDART00000004984
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr9_+_2762270 | 0.73 |
ENSDART00000123342
ENSDART00000001795 ENSDART00000177563 |
sp3a
|
sp3a transcription factor |
| chr4_+_43225728 | 0.73 |
ENSDART00000158639
ENSDART00000141339 ENSDART00000137770 ENSDART00000166019 |
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr18_+_47313715 | 0.72 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr6_-_41091151 | 0.71 |
ENSDART00000154963
ENSDART00000153818 |
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr24_-_23998897 | 0.71 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr8_-_30979494 | 0.71 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr6_+_48348415 | 0.71 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr19_-_12078583 | 0.70 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr4_+_60428812 | 0.70 |
ENSDART00000162049
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_+_357810 | 0.69 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr3_-_34816893 | 0.69 |
ENSDART00000084448
ENSDART00000154696 |
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr23_+_30967686 | 0.69 |
ENSDART00000144485
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr21_+_19525141 | 0.69 |
ENSDART00000058489
|
gzma
|
granzyme A |
| chr8_-_38403018 | 0.68 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr4_-_71471302 | 0.66 |
ENSDART00000187687
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr18_-_13360106 | 0.66 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr21_+_3901775 | 0.66 |
ENSDART00000053609
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr2_-_6519017 | 0.66 |
ENSDART00000181716
|
rgs1
|
regulator of G protein signaling 1 |
| chr8_+_5024468 | 0.66 |
ENSDART00000030938
|
adra1aa
|
adrenoceptor alpha 1Aa |
| chr13_+_12671513 | 0.65 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr3_+_32365811 | 0.65 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr24_-_40009446 | 0.64 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr20_+_53522059 | 0.64 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr24_+_19415124 | 0.63 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr7_-_7764287 | 0.63 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr4_+_71014655 | 0.62 |
ENSDART00000170837
|
si:dkeyp-80d11.14
|
si:dkeyp-80d11.14 |
| chr23_+_42336084 | 0.61 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr1_-_10640748 | 0.61 |
ENSDART00000103550
|
zgc:174945
|
zgc:174945 |
| chr17_+_51744450 | 0.61 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr8_+_39511932 | 0.61 |
ENSDART00000113511
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr5_+_24059598 | 0.60 |
ENSDART00000051547
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr12_+_18234557 | 0.60 |
ENSDART00000130741
|
fam20cb
|
family with sequence similarity 20, member Cb |
| chr17_+_32623931 | 0.59 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr1_+_57041549 | 0.59 |
ENSDART00000152198
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr23_-_36316352 | 0.59 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr1_+_46598502 | 0.58 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr18_-_37355666 | 0.58 |
ENSDART00000098914
|
yap1
|
Yes-associated protein 1 |
| chr4_-_71470455 | 0.58 |
ENSDART00000172685
|
si:ch211-76m11.5
|
si:ch211-76m11.5 |
| chr15_-_43978141 | 0.57 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr1_+_57176256 | 0.56 |
ENSDART00000152741
|
si:dkey-27j5.6
|
si:dkey-27j5.6 |
| chr4_+_69712223 | 0.56 |
ENSDART00000163035
ENSDART00000164475 |
si:dkey-238o14.1
zgc:174944
|
si:dkey-238o14.1 zgc:174944 |
| chr7_-_26125092 | 0.56 |
ENSDART00000079364
|
snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr18_+_47313899 | 0.56 |
ENSDART00000192389
ENSDART00000189592 ENSDART00000184281 |
barx2
|
BARX homeobox 2 |
| chr25_+_13205878 | 0.55 |
ENSDART00000162319
ENSDART00000162283 |
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr8_-_22288258 | 0.55 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr23_-_20361971 | 0.55 |
ENSDART00000133373
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr13_-_15982707 | 0.54 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr4_-_39487825 | 0.54 |
ENSDART00000161855
|
si:dkey-261o4.6
|
si:dkey-261o4.6 |
| chr10_-_22970723 | 0.54 |
ENSDART00000182145
ENSDART00000100802 |
FP102052.1
|
|
| chr4_-_56103062 | 0.53 |
ENSDART00000165784
ENSDART00000190846 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr21_+_43199237 | 0.53 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr23_+_42254960 | 0.53 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr3_-_32590164 | 0.53 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr19_+_4051535 | 0.52 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr2_+_11685742 | 0.51 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr4_-_52783184 | 0.51 |
ENSDART00000172283
|
si:dkey-4j21.2
|
si:dkey-4j21.2 |
| chr22_+_24603930 | 0.50 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr4_-_56673431 | 0.50 |
ENSDART00000161464
ENSDART00000191926 |
si:ch211-227p7.5
|
si:ch211-227p7.5 |
| chr7_-_26262978 | 0.50 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr4_-_39186893 | 0.50 |
ENSDART00000150371
ENSDART00000121804 ENSDART00000133901 |
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr4_-_33073382 | 0.50 |
ENSDART00000146414
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr20_-_26531850 | 0.50 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr4_-_34459615 | 0.50 |
ENSDART00000169144
ENSDART00000167022 |
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr4_+_70341246 | 0.49 |
ENSDART00000123408
|
si:ch211-76m11.3
|
si:ch211-76m11.3 |
| chr16_-_33816082 | 0.49 |
ENSDART00000181769
|
rspo1
|
R-spondin 1 |
| chr4_+_36616118 | 0.48 |
ENSDART00000160058
ENSDART00000170506 |
si:dkey-151g22.1
|
si:dkey-151g22.1 |
| chr4_-_38829098 | 0.48 |
ENSDART00000151932
|
si:dkey-59l11.10
|
si:dkey-59l11.10 |
| chr25_-_31907590 | 0.47 |
ENSDART00000149707
ENSDART00000149471 |
eif3ja
|
eukaryotic translation initiation factor 3, subunit Ja |
| chr12_+_46967789 | 0.46 |
ENSDART00000114866
|
oat
|
ornithine aminotransferase |
| chr5_-_49951106 | 0.46 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr4_+_70015788 | 0.46 |
ENSDART00000161133
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr4_-_34460301 | 0.46 |
ENSDART00000192977
|
si:ch211-64i20.3
|
si:ch211-64i20.3 |
| chr17_+_51743908 | 0.46 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr6_+_23935045 | 0.46 |
ENSDART00000162993
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr10_+_38593645 | 0.46 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
Network of associatons between targets according to the STRING database.
First level regulatory network of fosl2l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.7 | 2.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.5 | 3.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 1.3 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.3 | 1.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 1.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.3 | 0.8 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 1.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 1.0 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 1.5 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 3.1 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.2 | 0.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 1.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 2.2 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) arachidonate transport(GO:1903963) |
| 0.2 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.2 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 1.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.4 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.6 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 1.1 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.7 | GO:0045907 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 1.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.6 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 1.1 | GO:0016486 | peptide hormone processing(GO:0016486) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.4 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.4 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.8 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.3 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.8 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.1 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 4.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.8 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.0 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.4 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 2.6 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) actin filament network formation(GO:0051639) |
| 0.0 | 1.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.8 | GO:0048268 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) clathrin coat assembly(GO:0048268) |
| 0.0 | 1.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.3 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 2.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.6 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 2.3 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.6 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.1 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 1.2 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.8 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 1.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.7 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.6 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.7 | GO:0050922 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 0.8 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 1.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 3.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 2.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 29.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 1.3 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.4 | 2.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 1.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 1.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 1.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 0.7 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.2 | 1.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 2.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 1.9 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 3.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 2.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 2.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.7 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 11.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 5.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.4 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 1.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.8 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 1.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.0 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.8 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 7.4 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 3.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 5.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 3.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |