Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
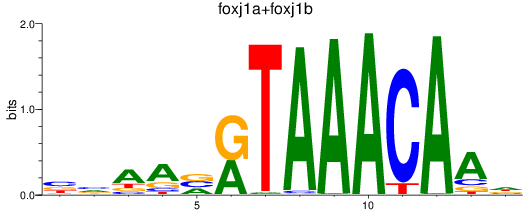
Results for foxj1a+foxj1b
Z-value: 1.02
Transcription factors associated with foxj1a+foxj1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj1b
|
ENSDARG00000088290 | forkhead box J1b |
|
foxj1a
|
ENSDARG00000101919 | forkhead box J1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj1a | dr11_v1_chr3_+_60716904_60716904 | -0.70 | 4.5e-15 | Click! |
| foxj1b | dr11_v1_chr12_+_20149305_20149311 | -0.68 | 3.1e-14 | Click! |
Activity profile of foxj1a+foxj1b motif
Sorted Z-values of foxj1a+foxj1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_7391400 | 13.72 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr4_+_7391110 | 12.60 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr7_+_22637515 | 10.96 |
ENSDART00000158698
|
si:dkey-112a7.5
|
si:dkey-112a7.5 |
| chr22_+_19553390 | 10.55 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr25_-_13188678 | 10.34 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr21_-_22951604 | 9.34 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr23_-_5683147 | 8.52 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr1_+_53945934 | 8.52 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr16_-_45235947 | 7.99 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr4_-_16353733 | 7.99 |
ENSDART00000186785
|
lum
|
lumican |
| chr4_-_12795436 | 7.69 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr25_+_19734038 | 7.19 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr6_-_55864687 | 7.14 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr18_+_35842933 | 6.89 |
ENSDART00000151587
ENSDART00000131121 |
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr15_+_34069746 | 6.83 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr17_-_6451801 | 6.66 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr14_-_36799280 | 6.57 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr19_+_18739085 | 6.50 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr4_-_12795030 | 6.45 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr21_-_5881344 | 6.42 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr14_-_35462148 | 6.32 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr12_-_8070969 | 6.18 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr8_-_30791266 | 6.13 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr22_-_28653074 | 6.11 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr4_-_6373735 | 6.01 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr25_+_8356707 | 5.95 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr8_+_10305400 | 5.82 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr12_+_17100021 | 5.79 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr17_-_23709347 | 5.74 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr1_+_36651059 | 5.49 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr19_-_24958393 | 5.46 |
ENSDART00000098592
|
si:ch211-195b13.6
|
si:ch211-195b13.6 |
| chr8_+_23034718 | 5.45 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr8_-_30791089 | 5.39 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr7_-_26270014 | 5.14 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr2_+_55365727 | 4.99 |
ENSDART00000162943
|
FP245456.1
|
|
| chr17_+_15297398 | 4.99 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr16_-_24195252 | 4.86 |
ENSDART00000136205
ENSDART00000048599 ENSDART00000161547 |
rps19
|
ribosomal protein S19 |
| chr1_-_26702930 | 4.84 |
ENSDART00000109297
ENSDART00000152389 |
foxe1
|
forkhead box E1 |
| chr24_+_9298198 | 4.79 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr25_-_13188214 | 4.74 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr18_-_48550426 | 4.73 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr24_-_37640705 | 4.69 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr14_+_15155684 | 4.67 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr13_-_31647323 | 4.62 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr20_+_15552657 | 4.61 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr24_-_31306724 | 4.59 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr21_+_10577527 | 4.56 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr4_+_5842433 | 4.54 |
ENSDART00000124085
ENSDART00000179848 |
usp18
|
ubiquitin specific peptidase 18 |
| chr21_-_3796461 | 4.54 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr7_-_28658143 | 4.45 |
ENSDART00000173556
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr17_-_27273296 | 4.36 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr23_-_1571682 | 4.33 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr22_-_37686966 | 4.25 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr11_-_6048490 | 4.15 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr21_-_11632403 | 4.03 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr25_-_13202208 | 3.97 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr12_-_11457625 | 3.96 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr7_-_28549361 | 3.94 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr24_+_37640626 | 3.87 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr13_-_43149063 | 3.84 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr17_-_20167206 | 3.83 |
ENSDART00000104874
ENSDART00000191995 |
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr19_+_30990815 | 3.79 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr11_-_183328 | 3.72 |
ENSDART00000168562
|
avil
|
advillin |
| chr25_+_37435720 | 3.69 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr2_+_38271392 | 3.67 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr14_+_26439227 | 3.60 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr12_+_25775734 | 3.59 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr16_+_33143503 | 3.58 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_+_6661343 | 3.53 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr20_+_25552057 | 3.51 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr14_+_30910114 | 3.48 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr5_-_34993242 | 3.48 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr23_+_7692042 | 3.41 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr23_+_10469955 | 3.35 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr6_-_37745508 | 3.25 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr15_-_25527580 | 3.24 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr3_+_29510818 | 3.22 |
ENSDART00000055407
ENSDART00000193743 ENSDART00000123619 |
rac2
|
Rac family small GTPase 2 |
| chr24_+_19542323 | 3.19 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr20_-_48172556 | 3.18 |
ENSDART00000097888
|
CABZ01059099.1
|
|
| chr3_+_32671146 | 3.15 |
ENSDART00000039466
|
rnf25
|
ring finger protein 25 |
| chr22_-_2959005 | 3.15 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr18_-_17399291 | 3.14 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr23_-_27123171 | 3.11 |
ENSDART00000023218
ENSDART00000146467 |
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr7_-_3912905 | 3.09 |
ENSDART00000143864
|
si:dkey-88n24.10
|
si:dkey-88n24.10 |
| chr7_-_28611145 | 3.08 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr20_+_26881600 | 3.07 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr5_-_1827575 | 3.07 |
ENSDART00000171193
|
mcc
|
mutated in colorectal cancers |
| chr10_-_3416258 | 3.03 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr23_+_25354856 | 3.02 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr2_+_21000334 | 3.00 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr15_-_17960228 | 2.96 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr17_-_26867725 | 2.91 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr20_+_53368611 | 2.89 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr23_-_17450746 | 2.88 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr9_-_11550711 | 2.85 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr18_-_46280578 | 2.84 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr20_+_6142433 | 2.84 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr10_+_41199660 | 2.82 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr7_-_30127082 | 2.80 |
ENSDART00000173749
|
alpk3b
|
alpha-kinase 3b |
| chr19_-_32600638 | 2.78 |
ENSDART00000143497
|
zgc:91944
|
zgc:91944 |
| chr24_-_19718077 | 2.78 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr5_-_66160415 | 2.76 |
ENSDART00000073895
|
mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr21_-_20840714 | 2.76 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr17_+_53284040 | 2.75 |
ENSDART00000165368
|
atxn3
|
ataxin 3 |
| chr16_-_26731928 | 2.71 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr16_+_26732086 | 2.69 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr3_-_13546610 | 2.67 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr25_+_37435335 | 2.64 |
ENSDART00000171602
|
chmp1a
|
charged multivesicular body protein 1A |
| chr1_+_9153141 | 2.63 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr7_-_24112484 | 2.63 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr10_-_42237304 | 2.61 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr11_+_18612421 | 2.61 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr17_+_25519089 | 2.58 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr19_-_46018152 | 2.57 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr13_-_21650404 | 2.49 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr3_-_40664868 | 2.49 |
ENSDART00000138783
ENSDART00000178567 |
rnf216
|
ring finger protein 216 |
| chr13_+_18533005 | 2.47 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr18_+_20226843 | 2.47 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr23_+_24598910 | 2.47 |
ENSDART00000126510
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr12_-_28537615 | 2.46 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr12_+_28117365 | 2.39 |
ENSDART00000066290
|
UTS2R
|
urotensin 2 receptor |
| chr15_-_44052927 | 2.38 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr2_+_19195841 | 2.37 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr19_+_33476557 | 2.33 |
ENSDART00000181800
|
triqk
|
triple QxxK/R motif containing |
| chr15_+_34675660 | 2.32 |
ENSDART00000009127
|
lta
|
lymphotoxin alpha (TNF superfamily, member 1) |
| chr23_-_45398622 | 2.32 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr3_-_36419641 | 2.31 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr5_+_12528693 | 2.29 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr2_+_19633493 | 2.27 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr7_+_46003449 | 2.24 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr6_-_43792179 | 2.22 |
ENSDART00000160849
|
foxp1b
|
forkhead box P1b |
| chr5_-_19014589 | 2.21 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr25_+_28555584 | 2.18 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr10_+_28428222 | 2.18 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr15_+_31886284 | 2.14 |
ENSDART00000156706
|
frya
|
furry homolog a (Drosophila) |
| chr16_+_22587661 | 2.13 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr10_+_40836378 | 2.12 |
ENSDART00000085792
|
trim69
|
tripartite motif containing 69 |
| chr23_-_31633201 | 2.10 |
ENSDART00000143335
ENSDART00000053531 |
slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr7_+_57108823 | 2.09 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr10_+_11260170 | 2.08 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr15_-_23647078 | 2.07 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr23_+_24085531 | 2.04 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr8_+_49570884 | 2.04 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr6_-_9695294 | 2.01 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr25_-_37435060 | 2.01 |
ENSDART00000102855
ENSDART00000148566 |
pdhx
|
pyruvate dehydrogenase complex, component X |
| chr14_+_16937997 | 1.99 |
ENSDART00000163013
ENSDART00000167856 |
limch1b
|
LIM and calponin homology domains 1b |
| chr6_+_51824596 | 1.98 |
ENSDART00000149003
|
rspo4
|
R-spondin 4 |
| chr10_+_22775253 | 1.97 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr14_-_28001986 | 1.95 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr13_-_7233811 | 1.95 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr7_+_4694924 | 1.93 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr3_+_25216790 | 1.92 |
ENSDART00000156544
|
c1qtnf6a
|
C1q and TNF related 6a |
| chr15_+_30158652 | 1.90 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr8_+_49065348 | 1.90 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr18_-_24988645 | 1.89 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr1_-_30510839 | 1.87 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr14_-_29799993 | 1.87 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr25_-_34740627 | 1.85 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr7_+_24165371 | 1.85 |
ENSDART00000173472
|
si:ch211-216p19.5
|
si:ch211-216p19.5 |
| chr15_-_28805493 | 1.84 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr22_+_37631234 | 1.80 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr2_+_19522082 | 1.79 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr7_-_13906409 | 1.78 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr4_-_17725008 | 1.78 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr19_+_619200 | 1.76 |
ENSDART00000050125
|
nupl2
|
nucleoporin like 2 |
| chr13_+_35635672 | 1.73 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr8_-_33154677 | 1.70 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr6_+_32834760 | 1.70 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr10_-_17501528 | 1.67 |
ENSDART00000144847
|
slc2a11l
|
solute carrier family 2 (facilitated glucose transporter), member 11-like |
| chr25_-_26893006 | 1.67 |
ENSDART00000006709
|
dldh
|
dihydrolipoamide dehydrogenase |
| chr17_+_24111392 | 1.65 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr22_+_11520249 | 1.64 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr20_-_26383368 | 1.64 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr3_+_46762703 | 1.63 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr2_-_19576640 | 1.62 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr10_+_23099890 | 1.62 |
ENSDART00000135890
|
LTN1
|
si:dkey-175g6.5 |
| chr2_-_17492080 | 1.62 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr11_+_25430851 | 1.61 |
ENSDART00000164999
ENSDART00000126403 |
si:dkey-13a21.4
|
si:dkey-13a21.4 |
| chr22_-_9714593 | 1.61 |
ENSDART00000188282
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr4_+_16725960 | 1.60 |
ENSDART00000034441
|
tcp11l2
|
t-complex 11, testis-specific-like 2 |
| chr19_+_4051535 | 1.60 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr21_+_3796620 | 1.60 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr16_-_7228276 | 1.59 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr1_+_11107688 | 1.59 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr7_-_60831082 | 1.59 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr8_-_22288258 | 1.56 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr10_-_14929392 | 1.56 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr23_+_38159715 | 1.56 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr10_-_35257458 | 1.56 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr17_+_6667441 | 1.56 |
ENSDART00000123503
ENSDART00000180381 ENSDART00000156140 |
smc6
|
structural maintenance of chromosomes 6 |
| chr6_-_18228358 | 1.54 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr20_-_2641233 | 1.51 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr6_+_40922572 | 1.50 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr20_-_18794789 | 1.49 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr17_+_43843536 | 1.49 |
ENSDART00000164097
|
FO704777.1
|
|
| chr21_-_11856143 | 1.47 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr19_-_10915898 | 1.47 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr7_+_4694762 | 1.46 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr7_-_50367326 | 1.43 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr9_+_11293830 | 1.43 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj1a+foxj1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 3.1 | 9.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 2.4 | 7.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 2.1 | 8.5 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 1.9 | 5.7 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.7 | 10.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 1.3 | 5.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 1.2 | 7.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 1.0 | 14.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.9 | 5.5 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.9 | 2.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.9 | 2.6 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.7 | 4.5 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.7 | 5.5 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.7 | 2.7 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.7 | 2.0 | GO:1901295 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.7 | 2.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.7 | 4.6 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.6 | 3.2 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.6 | 3.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.6 | 2.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.6 | 4.0 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.6 | 2.8 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.5 | 3.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.5 | 26.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.5 | 5.5 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.5 | 6.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.5 | 1.4 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.4 | 2.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.4 | 4.9 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.4 | 7.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.4 | 4.6 | GO:0045453 | bone resorption(GO:0045453) |
| 0.4 | 1.2 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.4 | 1.2 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.4 | 3.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.4 | 3.9 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.4 | 1.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 1.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 1.1 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.4 | 2.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 1.4 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.3 | 5.0 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.3 | 2.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 4.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.3 | 6.8 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.3 | 4.8 | GO:0003417 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.3 | 3.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 1.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 4.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.3 | 2.8 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.3 | 0.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.3 | 1.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.3 | 0.8 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.3 | 4.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.3 | 11.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.3 | 3.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.3 | 2.8 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.7 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 6.9 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 5.8 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 3.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.2 | 1.7 | GO:0046323 | glucose import(GO:0046323) |
| 0.2 | 1.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 8.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 4.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 0.8 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.6 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 1.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 2.8 | GO:0032094 | response to food(GO:0032094) |
| 0.2 | 2.9 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.2 | 3.1 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.2 | 2.3 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.2 | 4.3 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.2 | 1.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 3.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 4.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 5.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 0.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 2.0 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.2 | 1.6 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.2 | 2.1 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 2.3 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.7 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 3.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 2.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.4 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.4 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 2.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 2.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 3.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.6 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.5 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 2.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 7.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.5 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 1.6 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 3.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 2.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 4.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 2.4 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 1.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 2.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 2.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 3.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 4.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 1.9 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 1.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 0.5 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.7 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.9 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 1.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 1.5 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 2.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 1.0 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 2.3 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.9 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 4.8 | GO:0045597 | positive regulation of cell differentiation(GO:0045597) |
| 0.0 | 3.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 5.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 1.5 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 2.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 2.1 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 0.7 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.9 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 5.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 3.8 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.0 | 0.8 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.2 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.0 | 1.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 2.6 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.7 | GO:0006457 | protein folding(GO:0006457) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 14.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.0 | 2.9 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.9 | 14.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.8 | 34.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 1.8 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.6 | 3.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.5 | 2.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.5 | 6.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.4 | 1.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.4 | 2.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 2.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 2.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 2.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 2.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 5.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 5.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 1.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 2.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 4.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 2.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 3.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 2.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 3.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.7 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 6.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.9 | GO:0022627 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 3.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 0.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 18.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 8.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 8.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 1.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.6 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 4.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 4.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 8.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 54.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.8 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 5.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.9 | 5.7 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.8 | 7.1 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 1.0 | 5.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.9 | 5.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.8 | 5.5 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.8 | 8.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.7 | 2.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.7 | 2.8 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.7 | 3.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 4.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 4.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.5 | 1.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 2.0 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.5 | 1.4 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.4 | 2.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 1.2 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.4 | 10.5 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.4 | 2.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.4 | 1.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.3 | 1.0 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.3 | 11.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 4.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 3.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.3 | 2.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 2.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.3 | 4.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.6 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.2 | 2.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 2.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 1.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 2.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 10.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 3.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 2.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 3.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 3.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 2.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 4.9 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.5 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 1.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 3.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 1.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 2.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 12.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.9 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.5 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 2.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 3.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 15.0 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 4.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 4.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 3.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 6.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.7 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 2.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 3.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 25.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 11.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 3.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 4.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 6.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 2.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 1.2 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.3 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 2.3 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 2.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 14.1 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 5.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 8.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 3.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 2.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 5.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 3.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 3.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 2.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 5.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 6.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 8.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 6.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 14.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.7 | 8.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.7 | 4.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.5 | 4.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.5 | 4.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 2.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 7.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.3 | 5.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 7.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 1.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 3.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 11.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 5.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.1 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 3.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.4 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 4.0 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 2.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |