Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
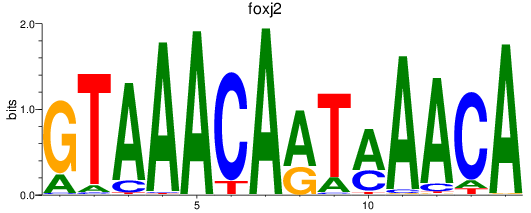
Results for foxj2
Z-value: 0.87
Transcription factors associated with foxj2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj2
|
ENSDARG00000057680 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj2 | dr11_v1_chr16_-_12784373_12784373 | 0.67 | 1.3e-13 | Click! |
Activity profile of foxj2 motif
Sorted Z-values of foxj2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_45177373 | 13.49 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr19_-_48340774 | 12.93 |
ENSDART00000168065
|
si:ch73-359m17.5
|
si:ch73-359m17.5 |
| chr14_+_20911310 | 8.21 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr8_+_32406885 | 8.16 |
ENSDART00000167600
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr15_+_42573909 | 8.10 |
ENSDART00000181801
|
CLDN8 (1 of many)
|
zgc:110333 |
| chr4_+_26357221 | 8.07 |
ENSDART00000187684
ENSDART00000101545 ENSDART00000148296 ENSDART00000171433 |
tnni1d
|
troponin I, skeletal, slow d |
| chr5_-_63302944 | 8.00 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr5_-_24201437 | 7.93 |
ENSDART00000114113
|
sox19a
|
SRY (sex determining region Y)-box 19a |
| chr15_-_29598444 | 6.48 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr9_-_6380653 | 6.15 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr18_-_44316920 | 6.05 |
ENSDART00000098599
|
si:ch211-151h10.2
|
si:ch211-151h10.2 |
| chr4_-_16353733 | 5.95 |
ENSDART00000186785
|
lum
|
lumican |
| chr10_-_15672862 | 5.77 |
ENSDART00000109231
|
mamdc2b
|
MAM domain containing 2b |
| chr4_+_76671012 | 5.22 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr11_+_37768298 | 5.12 |
ENSDART00000166886
|
sox13
|
SRY (sex determining region Y)-box 13 |
| chr8_-_46457233 | 4.50 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr16_+_35916371 | 4.38 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr15_-_9002155 | 4.34 |
ENSDART00000126708
|
rhoub
|
ras homolog family member Ub |
| chr22_-_28653074 | 4.34 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr18_-_15911394 | 4.27 |
ENSDART00000091339
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr12_-_44018667 | 4.24 |
ENSDART00000170692
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr19_+_18739085 | 4.07 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr18_+_35742838 | 3.91 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr16_+_44768361 | 3.55 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr7_-_28413224 | 3.48 |
ENSDART00000076502
|
rerglb
|
RERG/RAS-like b |
| chr18_+_25225524 | 3.37 |
ENSDART00000055563
|
si:dkeyp-59c12.1
|
si:dkeyp-59c12.1 |
| chr15_+_11883804 | 3.37 |
ENSDART00000163255
|
gpr184
|
G protein-coupled receptor 184 |
| chr23_-_27822920 | 3.35 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr14_+_21699129 | 3.30 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr8_-_39838660 | 3.24 |
ENSDART00000139266
|
ftr98
|
finTRIM family, member 98 |
| chr13_-_377256 | 3.18 |
ENSDART00000137398
|
ch1073-291c23.1
|
ch1073-291c23.1 |
| chr2_-_36918709 | 3.09 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr3_+_32112004 | 3.04 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr1_+_17527931 | 3.00 |
ENSDART00000131927
|
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr21_+_45757317 | 2.93 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr16_+_33143503 | 2.92 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_46277593 | 2.81 |
ENSDART00000045598
|
zgc:110626
|
zgc:110626 |
| chr25_-_35182347 | 2.79 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr19_+_19767567 | 2.78 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr7_-_28549361 | 2.76 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr23_-_17450746 | 2.64 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr20_+_34770197 | 2.54 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr6_-_14004772 | 2.54 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr14_-_5690363 | 2.52 |
ENSDART00000144539
|
tlx2
|
T cell leukemia homeobox 2 |
| chr7_-_43840680 | 2.51 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr19_-_1948236 | 2.42 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr6_+_7466223 | 2.33 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr3_-_57762247 | 2.23 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr15_-_23761580 | 2.23 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr12_-_17479078 | 2.21 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr19_+_6990970 | 2.18 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr4_-_12795436 | 2.14 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr15_-_4415917 | 2.12 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr1_-_49947290 | 2.09 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr14_+_21699414 | 2.07 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr25_+_29160102 | 2.02 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr14_-_34633960 | 2.00 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr18_+_7553950 | 1.88 |
ENSDART00000193420
ENSDART00000062143 |
zgc:77650
|
zgc:77650 |
| chr5_+_63302660 | 1.75 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr16_+_10422836 | 1.73 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr15_+_21254800 | 1.71 |
ENSDART00000142902
|
usf1
|
upstream transcription factor 1 |
| chr24_+_38201089 | 1.69 |
ENSDART00000132338
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr8_+_10305400 | 1.69 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr18_+_17534627 | 1.67 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr20_-_33966148 | 1.65 |
ENSDART00000148111
|
selp
|
selectin P |
| chr4_+_8168514 | 1.64 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr2_-_51507540 | 1.64 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr9_-_34842414 | 1.62 |
ENSDART00000126348
|
BX601644.1
|
Danio rerio cytokine receptor-like factor 2 (crlf2), mRNA. |
| chr4_-_19884440 | 1.62 |
ENSDART00000182979
ENSDART00000105967 |
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr5_-_69212184 | 1.59 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr4_-_12795030 | 1.58 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr19_+_20177887 | 1.57 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr25_-_13789955 | 1.54 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr1_-_54938137 | 1.54 |
ENSDART00000147212
|
crtac1a
|
cartilage acidic protein 1a |
| chr16_+_10318893 | 1.53 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr16_+_54210554 | 1.48 |
ENSDART00000172622
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr13_-_25476920 | 1.47 |
ENSDART00000131403
|
tial1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr17_-_53439866 | 1.46 |
ENSDART00000154826
|
mycbp
|
c-myc binding protein |
| chr17_+_51743908 | 1.35 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr2_+_39618951 | 1.35 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr8_-_43750062 | 1.35 |
ENSDART00000142243
|
ulk1a
|
unc-51 like autophagy activating kinase 1a |
| chr7_-_17337233 | 1.33 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr3_+_13862753 | 1.33 |
ENSDART00000168315
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr7_-_43840418 | 1.32 |
ENSDART00000174592
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr15_-_29162193 | 1.30 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr19_-_35492693 | 1.29 |
ENSDART00000135838
ENSDART00000051745 ENSDART00000177052 |
ptp4a2b
|
protein tyrosine phosphatase type IVA, member 2b |
| chr13_-_869808 | 1.29 |
ENSDART00000189029
|
AL929536.6
|
|
| chr6_-_29288155 | 1.28 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr17_+_33415319 | 1.27 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr16_+_13818500 | 1.27 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr4_-_19883985 | 1.26 |
ENSDART00000014440
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr15_-_28860282 | 1.25 |
ENSDART00000156738
|
gpr4
|
G protein-coupled receptor 4 |
| chr5_+_42957503 | 1.24 |
ENSDART00000192885
|
mob1ba
|
MOB kinase activator 1Ba |
| chr9_+_8942258 | 1.20 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr10_-_11261565 | 1.18 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr13_-_15986871 | 1.18 |
ENSDART00000189394
|
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr17_+_5985933 | 1.17 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr17_+_15534815 | 1.15 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr22_+_4442473 | 1.14 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr15_-_36357889 | 1.13 |
ENSDART00000156377
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr11_-_11878099 | 1.12 |
ENSDART00000188314
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr19_-_1920613 | 1.12 |
ENSDART00000186176
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr1_+_40566978 | 1.10 |
ENSDART00000137047
ENSDART00000135578 |
scoca
|
short coiled-coil protein a |
| chr18_+_41561285 | 1.09 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr17_+_33415542 | 1.09 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr3_-_55328548 | 1.08 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr13_+_30421472 | 1.07 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr4_-_31700186 | 1.03 |
ENSDART00000183986
ENSDART00000180890 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr6_-_18393476 | 1.03 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr8_-_10949847 | 1.02 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr21_+_38732945 | 1.00 |
ENSDART00000076157
|
rab24
|
RAB24, member RAS oncogene family |
| chr3_+_32129632 | 0.99 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr25_-_37465064 | 0.97 |
ENSDART00000186128
|
zgc:158366
|
zgc:158366 |
| chr18_+_41560822 | 0.97 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr2_+_22602301 | 0.96 |
ENSDART00000038514
|
sept2
|
septin 2 |
| chr10_+_15025006 | 0.94 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr2_-_38337122 | 0.93 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
| chr14_+_35279343 | 0.90 |
ENSDART00000187750
|
clint1a
|
clathrin interactor 1a |
| chr3_-_62194512 | 0.90 |
ENSDART00000074174
|
tbl3
|
transducin (beta)-like 3 |
| chr12_+_5977777 | 0.87 |
ENSDART00000152302
|
si:ch211-131k2.2
|
si:ch211-131k2.2 |
| chr16_-_41646164 | 0.86 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr3_+_34159192 | 0.85 |
ENSDART00000151119
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr25_+_17920361 | 0.85 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr6_+_11829867 | 0.85 |
ENSDART00000151044
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr4_-_75681004 | 0.84 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr25_-_3217115 | 0.84 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr4_+_13586455 | 0.81 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr19_+_48018802 | 0.80 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr12_-_8817999 | 0.80 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr6_+_12527725 | 0.80 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr7_-_20464468 | 0.79 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr24_+_17345521 | 0.78 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr18_+_20034023 | 0.78 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr4_+_14727212 | 0.76 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr4_+_43700319 | 0.76 |
ENSDART00000141967
|
si:ch211-226o13.1
|
si:ch211-226o13.1 |
| chr13_+_31687973 | 0.76 |
ENSDART00000076479
|
slc38a6
|
solute carrier family 38, member 6 |
| chr23_-_41762956 | 0.73 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr12_-_33706726 | 0.72 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr4_+_14727018 | 0.71 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr19_-_6134802 | 0.70 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr17_+_38602790 | 0.70 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr2_-_42492201 | 0.69 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr19_+_4856351 | 0.67 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr16_+_45922175 | 0.67 |
ENSDART00000018253
|
rbm8a
|
RNA binding motif protein 8A |
| chr10_+_42898103 | 0.66 |
ENSDART00000015872
|
zcchc9
|
zinc finger, CCHC domain containing 9 |
| chr3_-_31893008 | 0.65 |
ENSDART00000127297
|
ddx42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr23_+_20640875 | 0.63 |
ENSDART00000147382
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr16_-_5154024 | 0.63 |
ENSDART00000132069
ENSDART00000060635 |
dctn3
|
dynactin 3 (p22) |
| chr8_-_11202378 | 0.63 |
ENSDART00000147817
ENSDART00000174039 |
fam208b
|
family with sequence similarity 208, member B |
| chr23_-_10914275 | 0.63 |
ENSDART00000112965
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr22_-_718615 | 0.62 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr23_-_1571682 | 0.61 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr1_-_26444075 | 0.60 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr22_+_2254972 | 0.60 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr23_+_40275601 | 0.59 |
ENSDART00000076876
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr17_-_28797395 | 0.58 |
ENSDART00000134735
|
scfd1
|
sec1 family domain containing 1 |
| chr12_+_11352630 | 0.58 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr10_+_45031398 | 0.58 |
ENSDART00000160536
|
gnsb
|
glucosamine (N-acetyl)-6-sulfatase (Sanfilippo disease IIID), b |
| chr17_-_39761086 | 0.56 |
ENSDART00000032410
|
gpr132a
|
G protein-coupled receptor 132a |
| chr1_-_9940494 | 0.56 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr7_+_9904627 | 0.53 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr14_+_6991142 | 0.50 |
ENSDART00000157635
ENSDART00000059918 |
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr4_+_13586689 | 0.50 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr1_+_9994811 | 0.49 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr23_+_45966436 | 0.48 |
ENSDART00000172160
|
CABZ01069338.1
|
|
| chr17_-_681142 | 0.48 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr10_-_42776344 | 0.47 |
ENSDART00000190653
ENSDART00000130229 |
wdr45
|
WD repeat domain 45 |
| chr23_-_41762797 | 0.47 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr23_-_27607039 | 0.47 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr8_-_38477817 | 0.45 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr10_-_11261386 | 0.44 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr13_+_47007075 | 0.44 |
ENSDART00000109247
ENSDART00000183205 ENSDART00000180924 ENSDART00000133146 |
anapc1
|
anaphase promoting complex subunit 1 |
| chr25_+_3217419 | 0.44 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr23_+_40275400 | 0.43 |
ENSDART00000184259
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr10_+_36029537 | 0.43 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr18_+_38885309 | 0.43 |
ENSDART00000041597
|
arpp19a
|
cAMP-regulated phosphoprotein 19a |
| chr3_+_28860283 | 0.42 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr8_-_1278937 | 0.41 |
ENSDART00000112130
ENSDART00000179683 |
aaed1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr6_+_29288006 | 0.41 |
ENSDART00000043496
|
zgc:172121
|
zgc:172121 |
| chr19_+_46222428 | 0.40 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr19_-_11846958 | 0.40 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr15_+_618081 | 0.37 |
ENSDART00000181518
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr15_-_5901514 | 0.36 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr14_-_25577094 | 0.36 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr5_+_12590403 | 0.35 |
ENSDART00000133167
|
si:dkey-98f17.5
|
si:dkey-98f17.5 |
| chr3_+_59864872 | 0.34 |
ENSDART00000102014
|
mcrip1
|
MAPK regulated corepressor interacting protein 1 |
| chr25_+_17920668 | 0.34 |
ENSDART00000093358
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr9_-_32158288 | 0.33 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr16_-_20870143 | 0.32 |
ENSDART00000169541
ENSDART00000040727 |
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr11_-_45420212 | 0.32 |
ENSDART00000182042
ENSDART00000163185 |
ankrd13c
|
ankyrin repeat domain 13C |
| chr22_-_9728208 | 0.32 |
ENSDART00000185962
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr5_+_22970617 | 0.29 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr19_-_15229421 | 0.27 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr3_+_14571813 | 0.26 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr12_-_9790485 | 0.26 |
ENSDART00000027321
|
prdm9
|
PR domain containing 9 |
| chr18_+_3338228 | 0.25 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr2_-_29996036 | 0.24 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr13_-_10431476 | 0.23 |
ENSDART00000133968
|
camkmt
|
calmodulin-lysine N-methyltransferase |
| chr3_-_37588855 | 0.22 |
ENSDART00000149258
|
arf2a
|
ADP-ribosylation factor 2a |
| chr15_-_617797 | 0.22 |
ENSDART00000154224
|
si:ch73-144d13.7
|
si:ch73-144d13.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 2.1 | 6.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.1 | 5.4 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.0 | 3.0 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.9 | 8.2 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.7 | 2.2 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.5 | 4.3 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.4 | 2.2 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.4 | 2.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 1.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.4 | 2.5 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.4 | 1.8 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.3 | 1.0 | GO:0039531 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.3 | 3.3 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.3 | 0.9 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.3 | 1.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 1.7 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.3 | 3.7 | GO:0045824 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.3 | 8.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 0.9 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.9 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.7 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 1.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 8.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.5 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 6.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 1.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 4.1 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.8 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 1.7 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 3.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 6.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 2.1 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 3.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 1.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 1.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 1.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 2.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.8 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 1.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.9 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.1 | 1.7 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 1.2 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 2.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.4 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.1 | 2.2 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 3.9 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.3 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 4.2 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 3.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 2.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0048909 | anterior lateral line nerve development(GO:0048909) anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 1.8 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.4 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 3.8 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.3 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 1.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 4.3 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 2.9 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 2.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 1.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 1.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 3.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 5.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 8.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 2.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.7 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 2.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 4.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.9 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 10.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.9 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 6.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.6 | GO:0005912 | adherens junction(GO:0005912) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.7 | 2.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.6 | 3.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 4.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.5 | 2.3 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.4 | 1.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 8.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 2.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 2.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.3 | 0.9 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.2 | 0.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 0.9 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.2 | 8.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.3 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 3.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 3.4 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 1.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.5 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 7.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 2.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 4.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 14.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 6.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 3.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 8.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 6.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 4.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |