Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for foxk1_foxj3
Z-value: 0.50
Transcription factors associated with foxk1_foxj3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
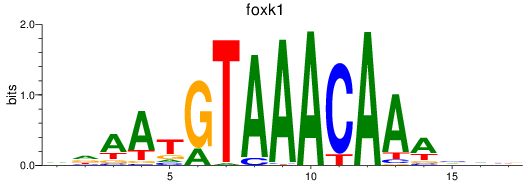
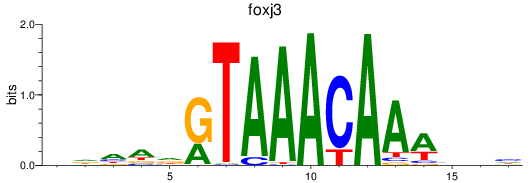
foxk1
|
ENSDARG00000037872 | forkhead box K1 |
|
foxj3
|
ENSDARG00000075774 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj3 | dr11_v1_chr11_-_26362294_26362334 | 0.48 | 9.4e-07 | Click! |
| foxk1 | dr11_v1_chr3_-_40933415_40933415 | 0.22 | 3.1e-02 | Click! |
Activity profile of foxk1_foxj3 motif
Sorted Z-values of foxk1_foxj3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_43025885 | 7.73 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr23_-_30787932 | 6.91 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr3_-_28258462 | 6.36 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr19_+_10396042 | 6.31 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr20_-_19590378 | 6.21 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr7_-_58098814 | 6.16 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr14_-_34044369 | 5.67 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_+_19207176 | 5.35 |
ENSDART00000087803
|
rln3a
|
relaxin 3a |
| chr11_+_6819050 | 4.93 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr2_-_11662851 | 4.23 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr12_+_25640480 | 4.17 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr15_+_8043751 | 4.07 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr1_+_16127825 | 4.07 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr6_-_32999646 | 3.90 |
ENSDART00000159510
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr13_+_28854438 | 3.50 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr19_-_28367413 | 3.47 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr24_-_21588914 | 3.45 |
ENSDART00000152054
|
gpr12
|
G protein-coupled receptor 12 |
| chr1_-_9277986 | 3.44 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr5_-_40190949 | 3.41 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr20_+_45741566 | 3.39 |
ENSDART00000113454
|
chgb
|
chromogranin B |
| chr19_-_10395683 | 3.35 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr25_-_11088839 | 3.34 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr21_-_28920245 | 3.33 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr1_-_44901163 | 3.13 |
ENSDART00000145354
|
tcirg1a
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3a |
| chr5_+_36611128 | 3.10 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr24_-_6678640 | 3.04 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr9_-_33785093 | 2.98 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr7_-_24828296 | 2.97 |
ENSDART00000138193
|
otub1b
|
OTU deubiquitinase, ubiquitin aldehyde binding 1b |
| chr5_+_49744713 | 2.93 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr14_+_36226243 | 2.78 |
ENSDART00000052569
|
pitx2
|
paired-like homeodomain 2 |
| chr7_+_22849657 | 2.74 |
ENSDART00000173483
|
pygmb
|
phosphorylase, glycogen, muscle b |
| chr18_+_6857071 | 2.74 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr12_-_29624638 | 2.70 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr9_+_41459759 | 2.68 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr20_-_19365875 | 2.66 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr19_-_19339285 | 2.57 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr2_+_16696052 | 2.55 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr23_-_28025943 | 2.52 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr6_-_30839763 | 2.51 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr1_+_45969240 | 2.50 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr3_+_34821327 | 2.49 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr14_-_17599452 | 2.42 |
ENSDART00000080042
|
rab33a
|
RAB33A, member RAS oncogene family |
| chr25_-_25142387 | 2.41 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr5_-_32323136 | 2.39 |
ENSDART00000110804
|
hspb15
|
heat shock protein, alpha-crystallin-related, b15 |
| chr11_+_23933016 | 2.39 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr16_+_43152727 | 2.29 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr19_-_5103313 | 2.27 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr16_+_6944564 | 2.26 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr12_+_5081759 | 2.21 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr24_-_6647275 | 2.17 |
ENSDART00000161494
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr5_-_38384289 | 2.05 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr13_+_22119798 | 2.04 |
ENSDART00000173206
ENSDART00000078652 ENSDART00000165842 |
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr13_+_29925397 | 2.04 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr15_+_19990068 | 2.03 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr12_+_19188542 | 2.03 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr5_-_66749535 | 2.01 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr5_-_23280098 | 2.00 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr16_+_25011994 | 1.99 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr16_-_26820634 | 1.98 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_+_25801648 | 1.97 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr20_-_26039841 | 1.96 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr1_-_22412042 | 1.96 |
ENSDART00000074678
|
chrnb3a
|
cholinergic receptor, nicotinic, beta polypeptide 3a |
| chr22_+_30335936 | 1.90 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr12_-_32066469 | 1.90 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr11_+_16152316 | 1.87 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr14_-_33095917 | 1.81 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr6_-_10305918 | 1.79 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr2_+_26179096 | 1.73 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr20_+_30445971 | 1.72 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr8_+_23093155 | 1.70 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr11_+_16153207 | 1.68 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr6_+_51932267 | 1.67 |
ENSDART00000156256
|
angpt4
|
angiopoietin 4 |
| chr2_-_30200206 | 1.65 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr25_-_24074500 | 1.64 |
ENSDART00000040410
|
th
|
tyrosine hydroxylase |
| chr5_-_45894802 | 1.64 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr21_-_27881752 | 1.61 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr18_+_5875268 | 1.61 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr2_-_8017579 | 1.61 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr16_+_28578352 | 1.61 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr3_-_60142530 | 1.60 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr8_-_18200003 | 1.60 |
ENSDART00000080014
|
rps8b
|
ribosomal protein S8b |
| chr18_+_17428506 | 1.55 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr19_+_2275019 | 1.55 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr13_-_27675212 | 1.55 |
ENSDART00000141035
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr6_+_38427570 | 1.54 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr16_-_17162843 | 1.53 |
ENSDART00000089386
|
iffo1b
|
intermediate filament family orphan 1b |
| chr11_-_8126223 | 1.53 |
ENSDART00000091617
ENSDART00000192391 ENSDART00000101561 |
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr6_+_38427357 | 1.52 |
ENSDART00000148678
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr19_-_5103141 | 1.51 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_-_16712111 | 1.50 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr11_-_21586157 | 1.49 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr24_-_7321928 | 1.48 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr8_-_39859688 | 1.48 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr12_+_19362335 | 1.48 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr22_-_18179214 | 1.48 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr3_-_28209001 | 1.47 |
ENSDART00000151178
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr19_+_46222918 | 1.46 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr3_-_51912019 | 1.44 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr22_-_29689981 | 1.44 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr25_+_13662606 | 1.40 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr13_+_38430466 | 1.40 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr14_-_48939560 | 1.39 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr20_-_18736281 | 1.38 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr15_+_26933196 | 1.37 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr14_-_30960470 | 1.36 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr14_-_26177156 | 1.34 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr22_+_4707663 | 1.33 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr3_-_26191960 | 1.33 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr11_+_25257022 | 1.31 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_+_46222428 | 1.31 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr20_-_16849306 | 1.31 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr5_-_19963537 | 1.29 |
ENSDART00000148146
|
si:dkey-234h16.7
|
si:dkey-234h16.7 |
| chr25_-_35360096 | 1.28 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr19_+_20724347 | 1.25 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr1_+_41886830 | 1.24 |
ENSDART00000185887
ENSDART00000084735 |
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr7_-_44963154 | 1.24 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr11_+_11120532 | 1.24 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr7_+_20110336 | 1.24 |
ENSDART00000179395
|
zgc:114045
|
zgc:114045 |
| chr5_-_54672763 | 1.23 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr16_+_6944311 | 1.22 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr11_-_13341483 | 1.21 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr20_+_46741074 | 1.19 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr13_+_29926094 | 1.19 |
ENSDART00000057528
|
cuedc2
|
CUE domain containing 2 |
| chr17_-_7218481 | 1.19 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr22_-_29689485 | 1.18 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr25_+_3677650 | 1.17 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr24_+_17007407 | 1.15 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr8_+_16990120 | 1.15 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr14_-_38843690 | 1.14 |
ENSDART00000183629
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr10_+_2529037 | 1.11 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr14_-_30490465 | 1.09 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr20_+_52774730 | 1.08 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr9_+_52411530 | 1.07 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr7_-_12909352 | 1.06 |
ENSDART00000172901
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr14_-_29799993 | 1.06 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr20_-_9428021 | 1.05 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr9_+_38163876 | 1.05 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr13_+_3667230 | 1.04 |
ENSDART00000131553
ENSDART00000189841 ENSDART00000183554 ENSDART00000018737 |
qkib
|
QKI, KH domain containing, RNA binding b |
| chr1_+_53321878 | 1.04 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr20_+_40457599 | 1.02 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr9_+_50600355 | 1.02 |
ENSDART00000187567
|
fign
|
fidgetin |
| chr2_+_5563077 | 1.01 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr21_-_43398122 | 0.98 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr17_+_24718272 | 0.98 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr1_-_12394048 | 0.98 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr25_-_13871118 | 0.97 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr2_-_24348642 | 0.97 |
ENSDART00000181739
|
ano8a
|
anoctamin 8a |
| chr16_-_30833519 | 0.96 |
ENSDART00000191321
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr14_-_17576391 | 0.95 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr5_-_12743196 | 0.95 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr20_-_23171430 | 0.94 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr17_-_14966384 | 0.94 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr21_+_8427059 | 0.93 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr7_+_52761841 | 0.92 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr18_+_26899316 | 0.92 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr13_+_9368621 | 0.92 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr21_+_30502002 | 0.91 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr18_-_13121983 | 0.91 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr25_-_25434479 | 0.91 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr13_-_21701323 | 0.90 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr5_-_51998708 | 0.90 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr2_-_24444838 | 0.90 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr25_+_18953575 | 0.89 |
ENSDART00000155610
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr15_+_39977461 | 0.87 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr25_-_16826219 | 0.87 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr17_-_35352690 | 0.86 |
ENSDART00000016053
|
rnf144aa
|
ring finger protein 144aa |
| chr12_+_32729470 | 0.86 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr4_-_14926637 | 0.86 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr16_-_49646625 | 0.84 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr15_+_5028608 | 0.83 |
ENSDART00000092809
|
abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr13_+_22280983 | 0.82 |
ENSDART00000173258
ENSDART00000173379 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr14_+_29581710 | 0.81 |
ENSDART00000188820
ENSDART00000193874 |
TENM2
|
si:dkey-34l15.2 |
| chr6_-_40922971 | 0.81 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr16_-_12097394 | 0.80 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr10_+_39084354 | 0.80 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr2_-_50372467 | 0.80 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr19_+_1878500 | 0.80 |
ENSDART00000113392
|
ggcta
|
gamma-glutamylcyclotransferase a |
| chr7_+_56472585 | 0.78 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr23_-_32304650 | 0.77 |
ENSDART00000143772
ENSDART00000085642 ENSDART00000188989 |
dgkaa
|
diacylglycerol kinase, alpha a |
| chr3_+_55031685 | 0.77 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr14_-_33177935 | 0.76 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr22_+_26798853 | 0.76 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr19_-_12965020 | 0.75 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr1_-_28749604 | 0.74 |
ENSDART00000148522
|
GK3P
|
zgc:172295 |
| chr18_-_20560007 | 0.74 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr17_+_24632440 | 0.73 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr22_+_38276024 | 0.72 |
ENSDART00000143792
|
rcor3
|
REST corepressor 3 |
| chr21_+_25071805 | 0.72 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr16_+_9580699 | 0.70 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_+_31951338 | 0.70 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr8_+_40081403 | 0.69 |
ENSDART00000138036
|
lrrc75ba
|
leucine rich repeat containing 75Ba |
| chr3_+_27786601 | 0.68 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr5_-_20446082 | 0.68 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr3_-_25369557 | 0.68 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr13_-_48431766 | 0.67 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr6_+_49021703 | 0.66 |
ENSDART00000149394
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr13_-_25842074 | 0.66 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr5_+_61799629 | 0.65 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr11_-_13341051 | 0.65 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr5_-_18046053 | 0.65 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxk1_foxj3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.2 | 6.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.9 | 3.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.9 | 2.7 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.8 | 2.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.8 | 2.4 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.6 | 1.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.6 | 3.4 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.6 | 2.8 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.5 | 1.6 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.5 | 2.0 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.5 | 2.0 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.5 | 1.9 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.4 | 1.3 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.4 | 1.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.4 | 2.0 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.4 | 5.7 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 2.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.3 | 2.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.3 | 1.5 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.3 | 1.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 2.6 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.3 | 1.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 12.4 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.2 | 1.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 4.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 1.2 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.2 | 3.1 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 0.8 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.2 | 1.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.8 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 0.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.2 | 1.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.2 | 2.4 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.2 | 2.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.2 | 3.5 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.2 | 2.0 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.2 | 0.8 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 0.9 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.4 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 1.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.5 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 4.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 1.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.9 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.6 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 1.9 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 2.0 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 2.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.3 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 1.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.5 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 2.3 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.1 | 3.9 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 3.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 1.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 1.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.2 | GO:0036315 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 1.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.0 | 1.0 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 4.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.5 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 3.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 2.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 7.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 1.0 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 1.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 2.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.3 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 3.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 1.0 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.4 | GO:0099565 | regulation of postsynaptic membrane potential(GO:0060078) excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 1.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.4 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 1.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.8 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 1.8 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 4.2 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 3.0 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 2.7 | GO:1904888 | cranial skeletal system development(GO:1904888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 3.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 3.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.4 | 6.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.4 | 5.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 1.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 3.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 3.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 1.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 4.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 3.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 9.7 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.2 | 0.9 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 0.6 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 2.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.8 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 2.0 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 3.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 4.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 1.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 8.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.0 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 2.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.0 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 3.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 2.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.7 | 3.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 3.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 2.7 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 2.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 1.9 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.4 | 5.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 6.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 2.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 3.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 2.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 0.9 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 3.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 0.8 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.6 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.2 | 2.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 3.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.3 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.2 | 0.5 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.2 | 4.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.9 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 1.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 5.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 3.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 2.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.7 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.8 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 2.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 5.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.4 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 8.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 2.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 2.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 2.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 5.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |