Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for foxm1
Z-value: 0.79
Transcription factors associated with foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxm1
|
ENSDARG00000003200 | forkhead box M1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxm1 | dr11_v1_chr4_-_5831522_5831522 | 0.32 | 1.7e-03 | Click! |
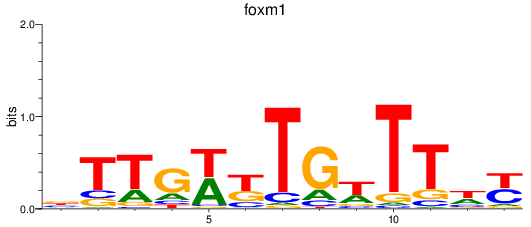
Activity profile of foxm1 motif
Sorted Z-values of foxm1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_5291012 | 5.62 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr25_-_22187397 | 4.82 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr16_+_21801277 | 4.46 |
ENSDART00000088407
|
trim108
|
tripartite motif containing 108 |
| chr3_-_19561058 | 4.26 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr2_+_11205795 | 4.17 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr12_+_17154655 | 4.17 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr24_+_20960216 | 4.00 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr18_+_17534627 | 3.41 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr5_-_24333684 | 3.22 |
ENSDART00000051553
|
znf703
|
zinc finger protein 703 |
| chr14_-_26177156 | 3.21 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr22_-_17671348 | 2.90 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr4_-_68913650 | 2.86 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr24_-_20915050 | 2.81 |
ENSDART00000133763
|
si:ch211-161h7.5
|
si:ch211-161h7.5 |
| chr15_-_23761580 | 2.80 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr24_+_17007407 | 2.75 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr2_-_29996036 | 2.64 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr14_+_20893065 | 2.63 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr19_-_340641 | 2.45 |
ENSDART00000183848
|
golph3l
|
golgi phosphoprotein 3-like |
| chr9_+_23770666 | 2.38 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr4_+_15944245 | 2.30 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr18_-_15932704 | 2.28 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr11_-_3646793 | 2.26 |
ENSDART00000077344
|
itih1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr5_+_31214341 | 2.25 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr14_-_22015232 | 2.18 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr14_+_31865099 | 2.17 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr9_-_38036984 | 2.16 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr14_+_45559268 | 2.15 |
ENSDART00000173152
|
RARRES3
|
zgc:154040 |
| chr1_+_59088205 | 2.13 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr7_+_26629084 | 2.02 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr3_-_37758487 | 2.02 |
ENSDART00000150938
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr7_+_15736230 | 2.01 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr13_-_22961605 | 1.95 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr20_-_39596338 | 1.93 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr20_-_22798794 | 1.90 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr21_+_37436907 | 1.90 |
ENSDART00000182611
ENSDART00000076328 |
pgrmc1
|
progesterone receptor membrane component 1 |
| chr6_+_40523370 | 1.88 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr5_-_22082918 | 1.87 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr14_-_8453192 | 1.84 |
ENSDART00000136947
|
eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr20_+_25904199 | 1.84 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr20_+_16881883 | 1.80 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr11_-_3647014 | 1.77 |
ENSDART00000171786
|
itih1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr6_+_40922572 | 1.77 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr5_-_67757188 | 1.76 |
ENSDART00000167168
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr4_-_12718367 | 1.75 |
ENSDART00000035259
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr10_+_22891126 | 1.72 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr3_+_1167026 | 1.69 |
ENSDART00000031823
ENSDART00000155340 |
triobpb
|
TRIO and F-actin binding protein b |
| chr25_-_24202576 | 1.68 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr6_-_14040136 | 1.65 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr22_-_9934854 | 1.63 |
ENSDART00000136404
|
si:dkey-253d23.11
|
si:dkey-253d23.11 |
| chr16_-_25841084 | 1.59 |
ENSDART00000159217
|
CR391937.1
|
|
| chr1_-_51038885 | 1.57 |
ENSDART00000035150
|
spast
|
spastin |
| chr12_-_9516981 | 1.52 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr4_-_890220 | 1.52 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr22_+_835728 | 1.51 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr5_-_9948497 | 1.51 |
ENSDART00000183196
|
tor4ab
|
torsin family 4, member Ab |
| chr1_+_52137528 | 1.47 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr2_-_39759059 | 1.45 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr1_+_58370526 | 1.43 |
ENSDART00000067775
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr15_+_37589698 | 1.43 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr21_-_13856689 | 1.41 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr14_-_33521071 | 1.41 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr15_-_41762530 | 1.39 |
ENSDART00000187125
ENSDART00000154971 |
ftr91
|
finTRIM family, member 91 |
| chr12_-_33558879 | 1.39 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr4_+_25558849 | 1.39 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr12_+_28888975 | 1.36 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr8_-_21071476 | 1.36 |
ENSDART00000184184
ENSDART00000100288 |
zgc:112962
|
zgc:112962 |
| chr14_+_45565891 | 1.35 |
ENSDART00000133389
ENSDART00000025549 |
zgc:92249
|
zgc:92249 |
| chr19_+_31904836 | 1.35 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr13_+_31550185 | 1.33 |
ENSDART00000127843
|
ppm1aa
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Aa |
| chr9_-_31108285 | 1.33 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr6_-_54444929 | 1.33 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr6_+_18569453 | 1.32 |
ENSDART00000171338
|
rhot1b
|
ras homolog family member T1 |
| chr2_+_58008980 | 1.32 |
ENSDART00000171264
|
si:ch211-155e24.3
|
si:ch211-155e24.3 |
| chr7_-_51102479 | 1.32 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr22_+_34615591 | 1.32 |
ENSDART00000189563
|
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr3_+_2719243 | 1.32 |
ENSDART00000189256
|
CR388047.3
|
|
| chr7_-_20836625 | 1.31 |
ENSDART00000192566
|
cldn15a
|
claudin 15a |
| chr9_-_38368138 | 1.30 |
ENSDART00000059574
|
ccdc93
|
coiled-coil domain containing 93 |
| chr13_+_33298338 | 1.29 |
ENSDART00000131892
ENSDART00000143895 |
iqcc
|
IQ motif containing C |
| chr18_+_27001115 | 1.26 |
ENSDART00000133547
|
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr6_+_56147812 | 1.25 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr4_-_43731342 | 1.25 |
ENSDART00000146627
|
si:ch211-226o13.3
|
si:ch211-226o13.3 |
| chr3_-_49514874 | 1.21 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr12_-_43428542 | 1.21 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr14_-_25577094 | 1.20 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr22_+_17606863 | 1.20 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| chr2_-_59231017 | 1.19 |
ENSDART00000143980
ENSDART00000184190 |
ftr29
|
finTRIM family, member 29 |
| chr4_-_22472653 | 1.17 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr5_-_51902935 | 1.17 |
ENSDART00000182160
|
mtx3
|
metaxin 3 |
| chr2_+_31838442 | 1.16 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr3_+_1107102 | 1.15 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr6_-_40922971 | 1.15 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr4_+_47445318 | 1.14 |
ENSDART00000183819
|
si:dkey-124l13.1
|
si:dkey-124l13.1 |
| chr20_+_9124369 | 1.14 |
ENSDART00000064150
|
si:ch211-59d15.9
|
si:ch211-59d15.9 |
| chr4_-_5691257 | 1.10 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr15_-_14210382 | 1.09 |
ENSDART00000179599
|
CR925813.1
|
|
| chr19_-_11966015 | 1.09 |
ENSDART00000123409
|
si:ch1073-296d18.1
|
si:ch1073-296d18.1 |
| chr16_+_40131473 | 1.09 |
ENSDART00000155421
ENSDART00000134732 ENSDART00000138699 |
cenpw
si:ch211-195p4.4
|
centromere protein W si:ch211-195p4.4 |
| chr11_-_18017918 | 1.08 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr18_+_18439332 | 1.08 |
ENSDART00000043843
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr6_+_35362225 | 1.07 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr1_+_55535827 | 1.07 |
ENSDART00000152784
|
adgre16
|
adhesion G protein-coupled receptor E16 |
| chr15_+_23534126 | 1.07 |
ENSDART00000152320
|
si:dkey-182i3.10
|
si:dkey-182i3.10 |
| chr11_-_42418374 | 1.06 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr21_-_23046606 | 1.06 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr17_-_50301313 | 1.06 |
ENSDART00000125772
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr10_+_8767541 | 1.06 |
ENSDART00000170272
|
itga1
|
integrin, alpha 1 |
| chr17_-_7818944 | 1.06 |
ENSDART00000135538
ENSDART00000037541 |
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr11_-_36474306 | 1.03 |
ENSDART00000170678
ENSDART00000123591 |
usp48
|
ubiquitin specific peptidase 48 |
| chr17_-_21162821 | 1.03 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr22_-_17606575 | 1.03 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr16_+_46410520 | 1.03 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr7_-_4125021 | 1.03 |
ENSDART00000167182
ENSDART00000173696 |
zgc:55733
|
zgc:55733 |
| chr4_+_75577480 | 1.02 |
ENSDART00000188196
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr12_+_30234209 | 1.02 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr4_-_23839789 | 1.02 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr8_+_40210398 | 1.01 |
ENSDART00000167612
|
rnf34a
|
ring finger protein 34a |
| chr7_+_60359347 | 1.00 |
ENSDART00000145201
ENSDART00000039827 |
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr19_-_46018152 | 1.00 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr3_-_33967767 | 1.00 |
ENSDART00000151493
ENSDART00000151160 |
ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr14_+_901847 | 0.98 |
ENSDART00000166991
|
si:ch73-208h1.2
|
si:ch73-208h1.2 |
| chr7_-_13906409 | 0.98 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr14_-_16082806 | 0.98 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr5_-_51903243 | 0.97 |
ENSDART00000125535
|
mtx3
|
metaxin 3 |
| chr16_+_25068576 | 0.97 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr18_+_44768829 | 0.96 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr13_+_5978809 | 0.95 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr3_-_54607166 | 0.95 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr8_-_22558773 | 0.94 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr7_+_39401388 | 0.94 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr15_+_47362728 | 0.93 |
ENSDART00000180712
|
CABZ01099833.1
|
|
| chr8_-_18211605 | 0.92 |
ENSDART00000114177
|
CT025742.1
|
|
| chr3_+_32714157 | 0.92 |
ENSDART00000131774
|
setd1a
|
SET domain containing 1A |
| chr4_-_12718103 | 0.91 |
ENSDART00000144388
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr7_+_4625915 | 0.90 |
ENSDART00000108557
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr13_+_44857087 | 0.89 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr4_+_60492313 | 0.89 |
ENSDART00000191043
ENSDART00000191631 |
si:dkey-211i20.2
|
si:dkey-211i20.2 |
| chr19_-_6134802 | 0.89 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr23_-_14990865 | 0.88 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr4_-_12388535 | 0.88 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr17_+_45639247 | 0.86 |
ENSDART00000153669
|
vcpkmt
|
valosin containing protein lysine (K) methyltransferase |
| chr5_-_38197080 | 0.86 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr4_+_67999732 | 0.85 |
ENSDART00000190597
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr4_+_75537772 | 0.85 |
ENSDART00000168380
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr4_+_29702703 | 0.84 |
ENSDART00000167771
|
si:ch211-214c20.1
|
si:ch211-214c20.1 |
| chr16_+_25809235 | 0.84 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr11_-_28614608 | 0.84 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr11_-_33618612 | 0.83 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr20_-_30938184 | 0.83 |
ENSDART00000147359
ENSDART00000062552 |
wtap
|
WT1 associated protein |
| chr4_-_60790123 | 0.82 |
ENSDART00000137702
|
si:dkey-254e13.6
|
si:dkey-254e13.6 |
| chr15_+_29393519 | 0.82 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr5_-_23596339 | 0.82 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr11_-_18107447 | 0.81 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr3_+_60589157 | 0.81 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr15_+_23784842 | 0.81 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr15_-_37589600 | 0.80 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr7_+_4572727 | 0.80 |
ENSDART00000137830
|
si:dkey-83f18.7
|
si:dkey-83f18.7 |
| chr5_+_50898849 | 0.79 |
ENSDART00000083317
|
arsk
|
arylsulfatase family, member K |
| chr22_+_5851122 | 0.79 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr15_-_23692359 | 0.76 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr19_-_7832439 | 0.75 |
ENSDART00000104703
ENSDART00000132336 |
znf687b
|
zinc finger protein 687b |
| chr18_+_27000850 | 0.75 |
ENSDART00000188906
ENSDART00000086131 |
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr16_+_53125918 | 0.74 |
ENSDART00000102170
|
CABZ01053976.1
|
|
| chr10_+_25272355 | 0.72 |
ENSDART00000140186
|
rab38b
|
RAB38b, member of RAS oncogene family |
| chr11_+_44502410 | 0.72 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr17_-_11439815 | 0.71 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr15_-_20710 | 0.69 |
ENSDART00000161218
|
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr15_-_44052927 | 0.67 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr2_-_10338759 | 0.65 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr1_+_12348213 | 0.65 |
ENSDART00000144920
ENSDART00000138759 ENSDART00000067082 |
clta
|
clathrin, light chain A |
| chr1_+_19538299 | 0.65 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr19_+_7759354 | 0.65 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr13_+_25421531 | 0.64 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr18_-_20458412 | 0.64 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr4_+_70171984 | 0.64 |
ENSDART00000158100
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr10_-_42237304 | 0.63 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr2_+_25840463 | 0.62 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr8_+_23147218 | 0.62 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr3_+_37112693 | 0.61 |
ENSDART00000055228
ENSDART00000144278 ENSDART00000138079 |
psmc3ip
|
PSMC3 interacting protein |
| chr18_-_20458840 | 0.60 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr22_+_2254972 | 0.60 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr10_+_26834985 | 0.60 |
ENSDART00000147518
|
fam89b
|
family with sequence similarity 89, member B |
| chr1_+_40566978 | 0.59 |
ENSDART00000137047
ENSDART00000135578 |
scoca
|
short coiled-coil protein a |
| chr1_-_23268569 | 0.59 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr21_-_22812417 | 0.58 |
ENSDART00000079151
ENSDART00000112175 |
trpc6a
|
transient receptor potential cation channel, subfamily C, member 6a |
| chr7_+_13995792 | 0.57 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr23_+_39611688 | 0.57 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr5_+_39099380 | 0.55 |
ENSDART00000166657
|
bmp2k
|
BMP2 inducible kinase |
| chr14_-_4177311 | 0.54 |
ENSDART00000128129
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr22_+_1734981 | 0.54 |
ENSDART00000158195
|
znf1159
|
zinc finger protein 1159 |
| chr19_+_18627100 | 0.54 |
ENSDART00000167245
|
vps52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr12_-_48566778 | 0.53 |
ENSDART00000063442
|
cyp4f3
|
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr3_-_37082618 | 0.53 |
ENSDART00000026701
ENSDART00000110716 |
tubg1
|
tubulin, gamma 1 |
| chr24_+_18948665 | 0.53 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr14_+_8638353 | 0.52 |
ENSDART00000163240
|
si:dkeyp-115e12.6
|
si:dkeyp-115e12.6 |
| chr6_-_29288155 | 0.52 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr6_-_19270484 | 0.52 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr4_-_36032177 | 0.52 |
ENSDART00000170143
|
zgc:174180
|
zgc:174180 |
| chr17_+_10593398 | 0.51 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr9_+_22780901 | 0.50 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxm1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.6 | 1.9 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.6 | 1.9 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.6 | 1.8 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.6 | 2.8 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.5 | 1.6 | GO:0051230 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.5 | 5.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.5 | 1.5 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.4 | 1.3 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.4 | 1.7 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.4 | 2.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.4 | 3.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.4 | 1.4 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.3 | 1.0 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.3 | 1.0 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.3 | 2.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 0.9 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.3 | 1.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 2.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.3 | 1.4 | GO:0031649 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 1.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.0 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.2 | 3.4 | GO:0010573 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.2 | 1.3 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 1.0 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.8 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 0.8 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.2 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 2.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 1.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 0.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 4.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.2 | 0.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 1.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.4 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 1.0 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.6 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 2.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.2 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.1 | 2.2 | GO:0010821 | regulation of mitochondrion organization(GO:0010821) |
| 0.1 | 1.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.4 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 1.3 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.8 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.1 | 0.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.6 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 2.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 2.6 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.1 | 3.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.3 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 1.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.3 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 0.0 | 1.1 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 6.1 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.5 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.1 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.5 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 1.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.9 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.6 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 3.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.5 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
| 0.0 | 0.2 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 1.1 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0003231 | cardiac ventricle development(GO:0003231) |
| 0.0 | 2.1 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 2.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.8 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.9 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 2.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 2.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.3 | 4.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.3 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.8 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 3.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 4.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.4 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 4.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 1.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.0 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 2.2 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.1 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 2.8 | GO:0005912 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.6 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.7 | 2.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 1.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.5 | 2.2 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 1.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.4 | 1.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.4 | 1.4 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 1.0 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.3 | 1.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 2.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 1.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 2.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 3.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.9 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 3.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 2.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 2.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 4.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 6.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0043022 | translation elongation factor activity(GO:0003746) ribosome binding(GO:0043022) |
| 0.0 | 0.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 4.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 2.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |