Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for foxn1
Z-value: 0.82
Transcription factors associated with foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxn1
|
ENSDARG00000011879 | forkhead box N1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxn1 | dr11_v1_chr15_-_28148314_28148314 | -0.12 | 2.5e-01 | Click! |
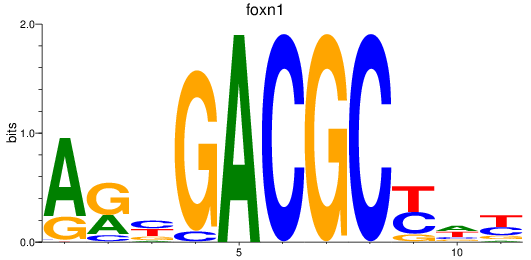
Activity profile of foxn1 motif
Sorted Z-values of foxn1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_19103490 | 5.79 |
ENSDART00000060561
|
csdc2a
|
cold shock domain containing C2, RNA binding a |
| chr13_+_51579851 | 5.37 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr23_+_19590006 | 4.92 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr7_+_23907692 | 4.80 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr3_-_35602233 | 4.73 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr3_+_33340939 | 4.62 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr24_-_29997145 | 4.41 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr19_-_21832441 | 4.39 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr14_+_8275115 | 4.29 |
ENSDART00000129055
|
nrg2b
|
neuregulin 2b |
| chr25_+_12640211 | 4.26 |
ENSDART00000165108
|
jph3
|
junctophilin 3 |
| chr1_+_19930520 | 4.23 |
ENSDART00000158344
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr10_+_21668606 | 4.16 |
ENSDART00000185751
|
CT737184.2
|
|
| chr4_+_2619132 | 4.03 |
ENSDART00000128807
|
gpr22a
|
G protein-coupled receptor 22a |
| chr8_+_6967108 | 3.95 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr17_-_46457622 | 3.81 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr12_+_26467847 | 3.81 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr17_-_45552602 | 3.74 |
ENSDART00000154844
ENSDART00000034432 |
susd4
|
sushi domain containing 4 |
| chr21_-_35325466 | 3.70 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr7_+_23495986 | 3.65 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr5_-_29643381 | 3.42 |
ENSDART00000034849
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr23_-_27345425 | 3.39 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr17_+_44780166 | 3.33 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr2_-_2096055 | 3.32 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr17_+_29345606 | 3.26 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr15_-_22147860 | 3.18 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr12_-_35988586 | 3.15 |
ENSDART00000157746
|
pde6gb
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog b |
| chr10_+_37500234 | 3.13 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr20_-_4738101 | 3.12 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr5_-_31901468 | 3.09 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr13_-_51247529 | 3.07 |
ENSDART00000191774
ENSDART00000083788 |
AL929217.1
|
|
| chr19_-_27966526 | 3.06 |
ENSDART00000141896
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr20_-_28884800 | 3.05 |
ENSDART00000134564
ENSDART00000132127 ENSDART00000135506 ENSDART00000075515 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr1_+_34203817 | 3.02 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr12_+_9703172 | 2.99 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr6_-_42377307 | 2.98 |
ENSDART00000129302
|
emc3
|
ER membrane protein complex subunit 3 |
| chr15_-_15449929 | 2.97 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr5_-_52216170 | 2.96 |
ENSDART00000158542
ENSDART00000192981 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr8_-_25846188 | 2.95 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr21_-_14251306 | 2.94 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr4_-_12102025 | 2.93 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr18_-_39787040 | 2.87 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr21_-_43606502 | 2.84 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr18_+_328689 | 2.83 |
ENSDART00000167841
|
ss18l2
|
synovial sarcoma translocation gene on chromosome 18-like 2 |
| chr16_+_50741154 | 2.74 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr1_-_21483832 | 2.73 |
ENSDART00000102790
|
glrba
|
glycine receptor, beta a |
| chr13_+_24287093 | 2.71 |
ENSDART00000058628
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr8_+_35172594 | 2.70 |
ENSDART00000177146
|
BX897670.1
|
|
| chr17_-_7861219 | 2.68 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr9_-_48296469 | 2.66 |
ENSDART00000058255
|
bbs5
|
Bardet-Biedl syndrome 5 |
| chr1_+_33969015 | 2.63 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr20_-_6196989 | 2.62 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr9_-_28399071 | 2.58 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr1_-_6494384 | 2.54 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr16_+_23531583 | 2.51 |
ENSDART00000146708
|
adar
|
adenosine deaminase, RNA-specific |
| chr11_+_6819050 | 2.47 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr17_+_37301860 | 2.45 |
ENSDART00000181531
ENSDART00000075978 |
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr15_+_15516612 | 2.38 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr2_+_42871831 | 2.36 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr14_-_2361692 | 2.32 |
ENSDART00000167696
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.4 |
| chr5_+_11812089 | 2.30 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr1_+_34295925 | 2.28 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr25_-_25142387 | 2.27 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr11_-_5563498 | 2.26 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr19_+_5135113 | 2.21 |
ENSDART00000151310
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr16_-_7362806 | 2.19 |
ENSDART00000166776
|
foxo6a
|
forkhead box O6 a |
| chr1_+_40237276 | 2.17 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr16_-_6205790 | 2.15 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr7_+_21859337 | 2.11 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr13_+_35925490 | 2.10 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr19_+_5134624 | 2.09 |
ENSDART00000151324
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr21_+_21279159 | 2.09 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr24_-_38374744 | 2.08 |
ENSDART00000007208
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr5_+_26075230 | 2.05 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr9_+_21793565 | 2.05 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr10_+_36695597 | 2.05 |
ENSDART00000169015
ENSDART00000171392 |
rab6a
|
RAB6A, member RAS oncogene family |
| chr4_-_8152746 | 2.03 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr1_+_31725154 | 1.99 |
ENSDART00000112333
ENSDART00000189801 |
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr2_-_16359042 | 1.96 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr23_-_10786400 | 1.92 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr23_+_23485858 | 1.88 |
ENSDART00000114067
|
agrn
|
agrin |
| chr19_-_27966780 | 1.87 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr11_-_38083397 | 1.85 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr21_+_3093419 | 1.84 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr11_+_23933016 | 1.82 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr2_+_37480669 | 1.78 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr16_-_31351419 | 1.77 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr4_-_1497384 | 1.74 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr7_+_22801465 | 1.72 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr20_-_31808779 | 1.71 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr7_-_12869545 | 1.70 |
ENSDART00000163045
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr6_-_37469775 | 1.68 |
ENSDART00000156546
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr12_-_13318944 | 1.68 |
ENSDART00000152201
ENSDART00000041394 |
emc9
|
ER membrane protein complex subunit 9 |
| chr11_-_22371105 | 1.67 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr6_+_39493864 | 1.66 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr19_+_26681848 | 1.64 |
ENSDART00000138322
|
si:dkey-27c15.3
|
si:dkey-27c15.3 |
| chr8_+_52530889 | 1.64 |
ENSDART00000127729
ENSDART00000170360 ENSDART00000162687 |
stambpb
|
STAM binding protein b |
| chr5_-_19932621 | 1.64 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr22_-_11614973 | 1.63 |
ENSDART00000063135
|
pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr8_-_43158486 | 1.62 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr13_+_18321140 | 1.62 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr3_-_32180796 | 1.61 |
ENSDART00000133191
ENSDART00000055308 |
pih1d1
|
PIH1 domain containing 1 |
| chr7_+_17782436 | 1.61 |
ENSDART00000173793
ENSDART00000165110 |
si:dkey-28a3.2
|
si:dkey-28a3.2 |
| chr9_-_24046287 | 1.60 |
ENSDART00000184313
|
ackr3a
|
atypical chemokine receptor 3a |
| chr1_-_45041272 | 1.60 |
ENSDART00000190615
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr3_+_34919810 | 1.57 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr15_-_23908779 | 1.56 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr23_-_35347714 | 1.55 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr5_-_52215926 | 1.55 |
ENSDART00000163973
ENSDART00000193602 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr21_+_21743599 | 1.54 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr11_-_19694334 | 1.53 |
ENSDART00000054735
|
SYNPR
|
si:dkey-30j16.3 |
| chr13_-_27916439 | 1.53 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr2_-_56131312 | 1.53 |
ENSDART00000097755
|
jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr5_+_1219187 | 1.48 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr1_+_41690402 | 1.47 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr5_-_54554583 | 1.45 |
ENSDART00000158865
ENSDART00000158069 |
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr21_+_1119046 | 1.43 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr12_-_10674606 | 1.43 |
ENSDART00000157919
|
med24
|
mediator complex subunit 24 |
| chr15_+_2421432 | 1.41 |
ENSDART00000193772
|
hephl1a
|
hephaestin-like 1a |
| chr16_+_26547152 | 1.41 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr13_-_21650404 | 1.39 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr24_+_19210001 | 1.38 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr2_+_16696052 | 1.37 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr17_+_10593398 | 1.36 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr8_-_24113575 | 1.36 |
ENSDART00000099692
ENSDART00000186211 |
dclre1b
|
DNA cross-link repair 1B |
| chr12_+_49100365 | 1.34 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr6_-_29612269 | 1.32 |
ENSDART00000104293
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr15_-_6976851 | 1.32 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr17_+_43032529 | 1.32 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr11_-_23687158 | 1.31 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr10_+_29137482 | 1.30 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr7_-_19146925 | 1.29 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr24_+_4373355 | 1.28 |
ENSDART00000179062
ENSDART00000093256 ENSDART00000138943 |
ccny
|
cyclin Y |
| chr8_+_668184 | 1.28 |
ENSDART00000183788
|
rnf165b
|
ring finger protein 165b |
| chr22_+_1786230 | 1.28 |
ENSDART00000169318
ENSDART00000164948 |
znf1154
|
zinc finger protein 1154 |
| chr3_-_22216771 | 1.27 |
ENSDART00000130546
|
maptb
|
microtubule-associated protein tau b |
| chr13_+_1439152 | 1.27 |
ENSDART00000159047
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr7_-_47850702 | 1.27 |
ENSDART00000109511
|
si:ch211-186j3.6
|
si:ch211-186j3.6 |
| chr18_+_19990412 | 1.24 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr16_+_32136550 | 1.23 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr4_-_73049281 | 1.22 |
ENSDART00000169165
|
LO018021.1
|
|
| chr15_-_40267485 | 1.22 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr12_-_4632519 | 1.20 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr7_+_24006875 | 1.19 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr21_-_14175838 | 1.18 |
ENSDART00000111659
|
whrna
|
whirlin a |
| chr18_-_5103931 | 1.18 |
ENSDART00000188091
|
pdcd10a
|
programmed cell death 10a |
| chr6_-_35106425 | 1.16 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr8_+_36500061 | 1.16 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr8_-_37043900 | 1.15 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr3_+_17951790 | 1.15 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr17_-_53022822 | 1.15 |
ENSDART00000103434
|
zgc:154061
|
zgc:154061 |
| chr8_-_18582922 | 1.15 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr10_-_43928937 | 1.15 |
ENSDART00000168014
ENSDART00000086220 |
mctp1b
|
multiple C2 domains, transmembrane 1b |
| chr15_+_2421729 | 1.14 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr6_+_51932267 | 1.14 |
ENSDART00000156256
|
angpt4
|
angiopoietin 4 |
| chr16_-_2390931 | 1.13 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr17_-_23895026 | 1.11 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr12_+_19320657 | 1.10 |
ENSDART00000100075
ENSDART00000066389 |
tmem184ba
|
transmembrane protein 184ba |
| chr6_+_20647155 | 1.09 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr15_+_19335646 | 1.09 |
ENSDART00000062569
|
acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr6_+_34038963 | 1.09 |
ENSDART00000057732
ENSDART00000192496 |
ap1m3
|
adaptor-related protein complex 1, mu 3 subunit |
| chr19_+_29303847 | 1.08 |
ENSDART00000009149
|
maco1a
|
macoilin 1a |
| chr1_-_50791280 | 1.06 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr10_-_36825984 | 1.05 |
ENSDART00000111104
|
phf12a
|
PHD finger protein 12a |
| chr8_+_18010978 | 1.03 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr4_+_76441200 | 1.02 |
ENSDART00000125751
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr20_+_52442870 | 1.01 |
ENSDART00000163100
|
arhgap39
|
Rho GTPase activating protein 39 |
| chr1_-_41982582 | 1.00 |
ENSDART00000014678
|
adra1d
|
adrenoceptor alpha 1D |
| chr8_-_21071711 | 1.00 |
ENSDART00000111600
ENSDART00000135204 ENSDART00000131371 ENSDART00000146532 ENSDART00000137606 |
zgc:112962
|
zgc:112962 |
| chr10_+_42374957 | 0.98 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr9_-_785444 | 0.98 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| chr18_-_16937008 | 0.95 |
ENSDART00000100117
ENSDART00000022640 ENSDART00000136541 |
znf143b
|
zinc finger protein 143b |
| chr6_+_33885828 | 0.92 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr6_+_10450000 | 0.92 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr13_+_19884631 | 0.90 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr6_-_45949121 | 0.90 |
ENSDART00000058555
|
tardbp
|
TAR DNA binding protein |
| chr6_+_6491013 | 0.87 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr24_-_18179535 | 0.87 |
ENSDART00000186112
|
cntnap2a
|
contactin associated protein like 2a |
| chr20_+_54333774 | 0.86 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr17_+_38476300 | 0.85 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr25_+_36405021 | 0.85 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr4_+_12013043 | 0.85 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr1_+_44523516 | 0.85 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr3_+_15773991 | 0.84 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr18_+_16715864 | 0.84 |
ENSDART00000079758
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr2_+_22531185 | 0.83 |
ENSDART00000171959
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr20_-_18915376 | 0.83 |
ENSDART00000063725
|
xkr6b
|
XK, Kell blood group complex subunit-related family, member 6b |
| chr19_+_10473612 | 0.81 |
ENSDART00000125718
|
si:ch211-171h4.7
|
si:ch211-171h4.7 |
| chr8_-_21071476 | 0.81 |
ENSDART00000184184
ENSDART00000100288 |
zgc:112962
|
zgc:112962 |
| chr22_-_6651382 | 0.81 |
ENSDART00000124308
|
si:ch211-209l18.4
|
si:ch211-209l18.4 |
| chr22_-_38459316 | 0.80 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr8_+_18010568 | 0.80 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr6_+_7977611 | 0.80 |
ENSDART00000112290
|
palm3
|
paralemmin 3 |
| chr5_+_23045096 | 0.80 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr6_+_59832786 | 0.79 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr9_-_2594410 | 0.78 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr8_-_5220125 | 0.76 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr5_+_36415978 | 0.76 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr9_-_41090048 | 0.75 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr7_-_29571615 | 0.75 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr5_+_35463688 | 0.75 |
ENSDART00000142525
ENSDART00000131286 |
erlin2
|
ER lipid raft associated 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxn1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) regulation of protein activation cascade(GO:2000257) |
| 1.0 | 2.9 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.8 | 3.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.8 | 2.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.8 | 3.2 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.7 | 2.1 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.7 | 2.7 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.6 | 3.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.6 | 1.9 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.6 | 1.8 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.6 | 2.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.6 | 4.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.5 | 2.7 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 3.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.5 | 2.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.5 | 1.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.5 | 2.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.5 | 2.8 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.4 | 6.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 4.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.4 | 3.0 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.4 | 1.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.4 | 2.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.4 | 1.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.4 | 1.1 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.4 | 1.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 5.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.3 | 2.6 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.3 | 1.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.3 | 1.4 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.3 | 1.3 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.3 | 0.8 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.2 | 1.6 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 1.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 0.9 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 1.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 0.9 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 3.0 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 2.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 1.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 3.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 2.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 2.0 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 0.9 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 4.6 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.2 | 0.8 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 4.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 1.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 4.0 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 1.0 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.1 | 1.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 2.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.0 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 4.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 6.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 2.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 2.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.5 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.5 | GO:0046329 | negative regulation of JUN kinase activity(GO:0043508) negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 2.3 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 0.6 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 3.0 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 5.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 0.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 1.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 1.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.3 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 1.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.2 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 0.7 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 1.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.9 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 1.7 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 1.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.6 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.2 | GO:1903573 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 2.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 3.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 4.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.6 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 2.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 2.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 3.3 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 4.8 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 1.6 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.7 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.3 | GO:0015908 | fatty acid transport(GO:0015908) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.7 | 2.7 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.5 | 3.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 1.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 4.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 6.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 1.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.4 | 1.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.4 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.3 | 2.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 1.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 2.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 1.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.3 | 6.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 4.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 3.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.2 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 2.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 3.1 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 3.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 2.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 4.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 3.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 1.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 8.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 4.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 1.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 2.4 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 2.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.9 | 2.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.8 | 4.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.8 | 3.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.7 | 6.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 1.2 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 3.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 1.1 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.3 | 3.0 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.3 | 2.7 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 2.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 4.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 3.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 2.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 4.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 1.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 1.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 4.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 3.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 2.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 3.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 3.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 2.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 4.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 2.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 1.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 4.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.5 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 2.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 4.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 3.0 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 2.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.6 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 5.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.1 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 4.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.1 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 3.8 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 4.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 2.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 2.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 8.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 5.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0051879 | Hsp70 protein binding(GO:0030544) Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.0 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.6 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 2.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 4.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.7 | 2.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 2.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 3.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.8 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 3.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |