Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
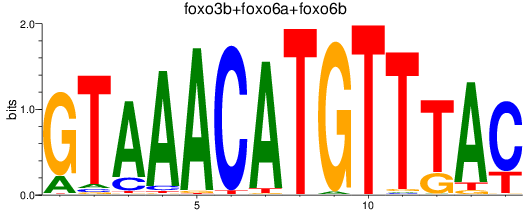
Results for foxo3b+foxo6a+foxo6b
Z-value: 0.41
Transcription factors associated with foxo3b+foxo6a+foxo6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo6b
|
ENSDARG00000024619 | forkhead box O6 b |
|
foxo3b
|
ENSDARG00000042904 | forkhead box O3b |
|
foxo6a
|
ENSDARG00000100486 | forkhead box O6 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo6a | dr11_v1_chr16_-_7362806_7362806 | 0.38 | 1.5e-04 | Click! |
| foxo6b | dr11_v1_chr19_+_15571290_15571290 | 0.36 | 4.3e-04 | Click! |
| foxo3b | dr11_v1_chr20_-_32332736_32332736 | 0.12 | 2.6e-01 | Click! |
Activity profile of foxo3b+foxo6a+foxo6b motif
Sorted Z-values of foxo3b+foxo6a+foxo6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_38125657 | 4.81 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr7_-_65492048 | 3.29 |
ENSDART00000162381
|
dkk3a
|
dickkopf WNT signaling pathway inhibitor 3a |
| chr21_+_11468934 | 2.60 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr19_-_13286722 | 1.92 |
ENSDART00000168296
ENSDART00000158330 |
zfpm2b
|
zinc finger protein, FOG family member 2b |
| chr16_-_29480335 | 1.86 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr24_+_32411753 | 1.86 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr13_+_36613359 | 1.81 |
ENSDART00000140598
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr11_+_41560792 | 1.81 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr6_+_58549080 | 1.80 |
ENSDART00000180117
|
stmn3
|
stathmin-like 3 |
| chr3_-_62380146 | 1.76 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr6_+_54221654 | 1.74 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr1_+_10018466 | 1.74 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr16_+_34479064 | 1.67 |
ENSDART00000159965
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr14_-_4682114 | 1.65 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr19_+_8481802 | 1.65 |
ENSDART00000050217
|
efna1a
|
ephrin-A1a |
| chr14_-_2221877 | 1.47 |
ENSDART00000106704
|
pcdh2ab1
|
protocadherin 2 alpha b 1 |
| chr19_-_19599372 | 1.46 |
ENSDART00000160002
ENSDART00000171664 |
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr18_-_40509006 | 1.46 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr21_-_43949208 | 1.46 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr7_-_30174882 | 1.42 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr15_-_12319065 | 1.42 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr24_+_32472155 | 1.40 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr3_-_48716422 | 1.36 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr20_+_25911342 | 1.30 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr7_+_30240791 | 1.29 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr5_+_28433215 | 1.29 |
ENSDART00000165072
ENSDART00000168005 |
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr10_-_21046917 | 1.25 |
ENSDART00000091004
|
pcdh1a
|
protocadherin 1a |
| chr17_-_28198099 | 1.23 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr16_-_25400257 | 1.16 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr9_-_785444 | 1.13 |
ENSDART00000012506
|
en1a
|
engrailed homeobox 1a |
| chr13_-_29420885 | 1.11 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr12_+_28799988 | 1.07 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr3_+_43460696 | 1.07 |
ENSDART00000164581
|
galr2b
|
galanin receptor 2b |
| chr3_+_62317990 | 1.07 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chr23_-_12158685 | 1.05 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr21_-_13123176 | 1.04 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr12_+_31713239 | 1.02 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr23_-_36670369 | 1.01 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr14_+_34547554 | 1.01 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr6_-_41138854 | 1.00 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr5_-_12031174 | 1.00 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr11_+_23933016 | 0.96 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr23_+_31405497 | 0.95 |
ENSDART00000053546
|
sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr7_-_36358735 | 0.92 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr14_-_32893785 | 0.92 |
ENSDART00000169380
|
slc25a43
|
solute carrier family 25, member 43 |
| chr5_+_38726854 | 0.91 |
ENSDART00000138484
ENSDART00000142867 |
si:dkey-58f10.7
|
si:dkey-58f10.7 |
| chr7_-_36358303 | 0.89 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr19_-_47323267 | 0.88 |
ENSDART00000190077
|
CU138512.1
|
|
| chr7_-_57949117 | 0.87 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr1_+_42874410 | 0.84 |
ENSDART00000153506
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr8_+_47897734 | 0.81 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr18_+_10689772 | 0.79 |
ENSDART00000126441
|
lepa
|
leptin a |
| chr1_+_31942961 | 0.77 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr2_+_10134345 | 0.75 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr24_-_6345965 | 0.75 |
ENSDART00000178287
|
zgc:174877
|
zgc:174877 |
| chr12_+_11352630 | 0.74 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr4_-_71110826 | 0.74 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr2_-_43850897 | 0.73 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr14_-_46238186 | 0.73 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr9_-_16877456 | 0.73 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr16_+_46000956 | 0.73 |
ENSDART00000101753
ENSDART00000162393 |
mtmr11
|
myotubularin related protein 11 |
| chr9_+_38624478 | 0.71 |
ENSDART00000077389
ENSDART00000114728 |
znf148
|
zinc finger protein 148 |
| chr3_-_26341959 | 0.69 |
ENSDART00000169344
ENSDART00000142878 ENSDART00000087196 |
zgc:153240
|
zgc:153240 |
| chr22_-_21676364 | 0.67 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr6_+_28124393 | 0.63 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr15_-_8177919 | 0.61 |
ENSDART00000101463
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr19_-_23470709 | 0.61 |
ENSDART00000138884
|
pimr80
|
Pim proto-oncogene, serine/threonine kinase, related 80 |
| chr9_-_3834686 | 0.59 |
ENSDART00000141492
|
myo3b
|
myosin IIIB |
| chr25_-_16832705 | 0.59 |
ENSDART00000171336
ENSDART00000189396 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr21_-_37973081 | 0.58 |
ENSDART00000136569
|
ripply1
|
ripply transcriptional repressor 1 |
| chr2_-_7131657 | 0.57 |
ENSDART00000175565
ENSDART00000092116 |
extl2
|
exostosin-like glycosyltransferase 2 |
| chr5_+_57320113 | 0.57 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr6_+_18531932 | 0.57 |
ENSDART00000165271
|
suz12b
|
SUZ12 polycomb repressive complex 2b subunit |
| chr22_-_34995333 | 0.56 |
ENSDART00000110900
|
kcnip2
|
Kv channel interacting protein 2 |
| chr22_+_2524615 | 0.56 |
ENSDART00000134277
|
znf1003
|
zinc finger protein 1003 |
| chr9_-_3149896 | 0.56 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr18_-_20560007 | 0.56 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr22_+_2533514 | 0.56 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr17_-_50436199 | 0.56 |
ENSDART00000156494
|
si:ch211-235i11.6
|
si:ch211-235i11.6 |
| chr24_-_6345647 | 0.55 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr7_+_61906903 | 0.53 |
ENSDART00000108540
|
tdrd7b
|
tudor domain containing 7 b |
| chr12_+_46582168 | 0.53 |
ENSDART00000189402
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr16_-_21181128 | 0.52 |
ENSDART00000133403
|
pde11al
|
phosphodiesterase 11a, like |
| chr13_-_8601184 | 0.52 |
ENSDART00000045086
|
prkceb
|
protein kinase C, epsilon b |
| chr22_+_7486867 | 0.52 |
ENSDART00000034586
|
CELA1 (1 of many)
|
zgc:112302 |
| chr3_-_31078348 | 0.52 |
ENSDART00000055360
|
elob
|
elongin B |
| chr5_-_26118855 | 0.51 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr21_-_29110607 | 0.51 |
ENSDART00000181595
|
havcr1
|
hepatitis A virus cellular receptor 1 |
| chr21_+_22828500 | 0.51 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr18_+_48943875 | 0.50 |
ENSDART00000076803
|
FO681288.1
|
|
| chr14_-_17305849 | 0.50 |
ENSDART00000160971
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr1_-_10347198 | 0.49 |
ENSDART00000109141
ENSDART00000152565 |
si:ch211-243g6.3
|
si:ch211-243g6.3 |
| chr10_-_25570647 | 0.48 |
ENSDART00000134215
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr6_+_15762647 | 0.48 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr21_+_17301790 | 0.48 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr6_-_18531760 | 0.48 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr22_+_7462997 | 0.46 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr5_+_29652198 | 0.46 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr19_-_21766461 | 0.45 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr6_+_55819038 | 0.45 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr14_-_17305657 | 0.44 |
ENSDART00000168853
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr2_-_44199722 | 0.43 |
ENSDART00000140633
ENSDART00000145728 |
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr17_-_50454957 | 0.43 |
ENSDART00000171616
ENSDART00000075195 |
si:ch211-235i11.7
|
si:ch211-235i11.7 |
| chr6_-_46560207 | 0.42 |
ENSDART00000187607
|
kcnb1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr22_+_7480465 | 0.42 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr13_-_32626247 | 0.42 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr6_-_51386656 | 0.42 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr6_+_10077756 | 0.42 |
ENSDART00000150982
|
htr2ab
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate b |
| chr11_+_25044082 | 0.41 |
ENSDART00000123263
|
phf20a
|
PHD finger protein 20, a |
| chr18_+_41527877 | 0.41 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr5_-_31035198 | 0.40 |
ENSDART00000086534
|
cyb5d2
|
cytochrome b5 domain containing 2 |
| chr1_-_45752010 | 0.40 |
ENSDART00000101410
|
si:ch211-214c7.5
|
si:ch211-214c7.5 |
| chr14_-_6402769 | 0.40 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr7_+_32021669 | 0.39 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr4_-_64123545 | 0.39 |
ENSDART00000170040
|
BX914205.2
|
|
| chr16_-_31351419 | 0.39 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr20_+_22067337 | 0.38 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr19_+_19600297 | 0.38 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr4_-_77377596 | 0.37 |
ENSDART00000186068
|
slco1e1
|
solute carrier organic anion transporter family, member 1E1 |
| chr23_+_17878969 | 0.36 |
ENSDART00000154934
ENSDART00000154140 ENSDART00000156331 |
si:ch73-390p7.2
|
si:ch73-390p7.2 |
| chr5_-_30145939 | 0.36 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr25_+_28158352 | 0.35 |
ENSDART00000151854
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr2_-_31540922 | 0.33 |
ENSDART00000056686
|
mrc1b
|
mannose receptor, C type 1b |
| chr4_-_12781182 | 0.33 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr1_+_6357059 | 0.32 |
ENSDART00000166620
|
hecw2b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2b |
| chr7_-_19966406 | 0.32 |
ENSDART00000010932
|
snx15
|
sorting nexin 15 |
| chr8_-_16464453 | 0.32 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr8_-_36125849 | 0.31 |
ENSDART00000159581
|
CT583723.1
|
|
| chr4_+_75458537 | 0.31 |
ENSDART00000166278
|
si:ch211-266c8.1
|
si:ch211-266c8.1 |
| chr8_-_14179798 | 0.31 |
ENSDART00000040645
ENSDART00000146749 |
rhoaa
|
ras homolog gene family, member Aa |
| chr1_+_19708508 | 0.31 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr16_-_8663493 | 0.30 |
ENSDART00000014944
ENSDART00000181533 ENSDART00000182721 ENSDART00000162988 |
cobl
|
cordon-bleu WH2 repeat protein |
| chr5_-_25733745 | 0.30 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr6_+_52859495 | 0.29 |
ENSDART00000103135
|
or134-1
|
odorant receptor, family H, subfamily 134, member 1 |
| chr2_-_43850637 | 0.29 |
ENSDART00000136077
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr17_-_5610514 | 0.29 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr17_-_50472864 | 0.29 |
ENSDART00000161066
ENSDART00000156963 |
si:ch211-235i11.3
|
si:ch211-235i11.3 |
| chr4_+_20536120 | 0.28 |
ENSDART00000125685
|
zmp:0000000650
|
zmp:0000000650 |
| chr18_-_46208581 | 0.28 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr14_-_17306261 | 0.28 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr2_+_24317465 | 0.27 |
ENSDART00000139339
ENSDART00000136284 |
mrpl34
|
mitochondrial ribosomal protein L34 |
| chr1_+_5576151 | 0.27 |
ENSDART00000109756
|
cpo
|
carboxypeptidase O |
| chr12_+_17603528 | 0.27 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr10_+_38526496 | 0.26 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr2_-_24317240 | 0.26 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr22_-_10440688 | 0.26 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr22_-_5958066 | 0.26 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr24_+_19591893 | 0.26 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr22_-_23590069 | 0.25 |
ENSDART00000172067
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr22_-_17499513 | 0.25 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr2_+_42318012 | 0.24 |
ENSDART00000138137
|
ftr08
|
finTRIM family, member 8 |
| chr1_+_8314826 | 0.24 |
ENSDART00000160440
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr17_+_8175998 | 0.23 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr6_+_53429228 | 0.23 |
ENSDART00000165067
|
abhd14b
|
abhydrolase domain containing 14B |
| chr23_+_4777612 | 0.23 |
ENSDART00000135933
|
vgll4a
|
vestigial-like family member 4a |
| chr6_+_30091811 | 0.21 |
ENSDART00000088403
|
meltf
|
melanotransferrin |
| chr15_-_44465679 | 0.21 |
ENSDART00000054605
|
aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr3_+_52684556 | 0.21 |
ENSDART00000139037
|
pgls
|
6-phosphogluconolactonase |
| chr1_+_6646529 | 0.20 |
ENSDART00000144641
ENSDART00000103701 ENSDART00000138919 |
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr3_-_46115232 | 0.20 |
ENSDART00000074537
|
shisa9b
|
shisa family member 9b |
| chr4_+_42604252 | 0.19 |
ENSDART00000184850
|
CR925768.1
|
|
| chr21_+_17541634 | 0.18 |
ENSDART00000191700
ENSDART00000163238 |
stom
|
stomatin |
| chr20_-_7069612 | 0.18 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr13_-_31377299 | 0.17 |
ENSDART00000026692
|
ubtd1a
|
ubiquitin domain containing 1a |
| chr7_-_13362590 | 0.17 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr8_-_53044089 | 0.16 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr17_-_2834764 | 0.16 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr3_+_34180835 | 0.16 |
ENSDART00000055252
|
timm29
|
translocase of inner mitochondrial membrane 29 |
| chr10_+_25369254 | 0.16 |
ENSDART00000164375
|
bach1b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 b |
| chr21_-_17037907 | 0.15 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr18_-_3103827 | 0.15 |
ENSDART00000165048
|
aamdc
|
adipogenesis associated, Mth938 domain containing |
| chr15_+_791220 | 0.15 |
ENSDART00000136137
|
si:dkey-7i4.1
|
si:dkey-7i4.1 |
| chr6_+_3516926 | 0.15 |
ENSDART00000151607
|
faah
|
fatty acid amide hydrolase |
| chr11_-_23697217 | 0.15 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_+_23130389 | 0.15 |
ENSDART00000142834
|
si:dkey-218h11.6
|
si:dkey-218h11.6 |
| chr18_+_33100606 | 0.14 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr6_+_46259950 | 0.14 |
ENSDART00000032326
|
zgc:162324
|
zgc:162324 |
| chr15_-_31406093 | 0.14 |
ENSDART00000123444
|
or111-8
|
odorant receptor, family D, subfamily 111, member 8 |
| chr25_+_4855549 | 0.13 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr2_-_24907741 | 0.13 |
ENSDART00000155013
|
si:dkey-149i17.11
|
si:dkey-149i17.11 |
| chr13_-_25842074 | 0.13 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr15_-_8191992 | 0.11 |
ENSDART00000155381
ENSDART00000190714 |
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr22_-_34715364 | 0.11 |
ENSDART00000189348
|
CU539054.1
|
|
| chr5_+_56119975 | 0.11 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr16_+_46410520 | 0.11 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr12_-_28799642 | 0.09 |
ENSDART00000066303
|
mrpl10
|
mitochondrial ribosomal protein L10 |
| chr4_-_53047883 | 0.09 |
ENSDART00000157496
|
si:dkey-201g16.5
|
si:dkey-201g16.5 |
| chr22_+_18319230 | 0.08 |
ENSDART00000184747
ENSDART00000184649 |
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr5_-_68795063 | 0.08 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr5_-_51998708 | 0.08 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr5_+_29652513 | 0.08 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr18_-_44526940 | 0.07 |
ENSDART00000077125
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr23_+_17417539 | 0.07 |
ENSDART00000182605
|
BX649300.2
|
|
| chr2_-_51388214 | 0.07 |
ENSDART00000163838
|
si:dkeyp-104b3.18
|
si:dkeyp-104b3.18 |
| chr18_-_10995410 | 0.06 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr8_-_53044300 | 0.06 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr20_+_33994580 | 0.05 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr11_+_45422206 | 0.05 |
ENSDART00000182548
|
hrasls
|
HRAS-like suppressor |
| chr7_-_30932094 | 0.05 |
ENSDART00000057677
|
terb2
|
telomere repeat binding bouquet formation protein 2 |
| chr3_+_33918510 | 0.05 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo3b+foxo6a+foxo6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 1.1 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.4 | 1.1 | GO:0042822 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 1.4 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.3 | 1.0 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 1.0 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 2.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 1.5 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.8 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 2.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.5 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 1.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.0 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.7 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.2 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 1.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 3.0 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.8 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.4 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.4 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 1.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 3.3 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.0 | GO:0044821 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.6 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0021591 | ventricular system development(GO:0021591) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 1.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 2.6 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.4 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 2.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 3.3 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.3 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.6 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.0 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 9.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.5 | 1.8 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.4 | 1.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 1.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.3 | 1.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 2.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 1.8 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 1.6 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 1.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 1.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |