Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
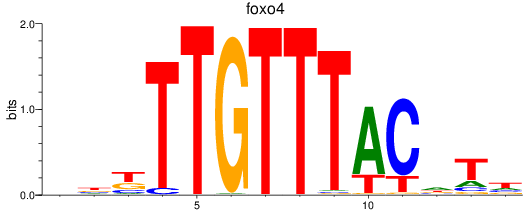
Results for foxo4
Z-value: 1.14
Transcription factors associated with foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo4
|
ENSDARG00000055792 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo4 | dr11_v1_chr14_+_30910114_30910114 | 0.57 | 2.0e-09 | Click! |
Activity profile of foxo4 motif
Sorted Z-values of foxo4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_36378494 | 13.31 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr13_+_22675802 | 10.43 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr7_+_22637515 | 8.78 |
ENSDART00000158698
|
si:dkey-112a7.5
|
si:dkey-112a7.5 |
| chr5_+_37837245 | 8.43 |
ENSDART00000171617
|
epd
|
ependymin |
| chr12_+_25640480 | 7.42 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr6_-_30839763 | 7.05 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr5_+_28271412 | 7.01 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr20_-_39273987 | 6.92 |
ENSDART00000127173
|
clu
|
clusterin |
| chr14_-_34044369 | 6.77 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr4_-_4256300 | 6.66 |
ENSDART00000103319
ENSDART00000150279 |
cd9b
|
CD9 molecule b |
| chr24_+_17007407 | 6.43 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr18_-_17485419 | 6.03 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr6_+_48618512 | 5.77 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr11_+_18873619 | 5.73 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr12_+_19188542 | 5.73 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr12_-_11457625 | 5.70 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr20_-_39273505 | 5.69 |
ENSDART00000153114
|
clu
|
clusterin |
| chr7_+_60551133 | 5.54 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr11_+_25112269 | 5.36 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr18_+_17428506 | 5.12 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr9_+_9502610 | 5.03 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr19_-_5103141 | 5.02 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr10_+_33744098 | 4.92 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr14_-_7885707 | 4.87 |
ENSDART00000029981
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr7_-_30082931 | 4.69 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr1_-_37383539 | 4.69 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr16_-_43025885 | 4.68 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr13_-_25842074 | 4.64 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr13_+_22280983 | 4.59 |
ENSDART00000173258
ENSDART00000173379 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr22_-_29689981 | 4.58 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr22_+_4707663 | 4.58 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr19_-_5103313 | 4.57 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr6_+_36942966 | 4.35 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr9_+_38163876 | 4.34 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr6_-_40657653 | 4.34 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr12_+_27213733 | 4.33 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr23_-_36449111 | 4.28 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr16_-_15988320 | 4.27 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr9_+_3388099 | 4.23 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr3_+_32403758 | 4.23 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr22_-_29689485 | 4.23 |
ENSDART00000182173
|
pdcd4b
|
programmed cell death 4b |
| chr21_+_25071805 | 4.18 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr16_+_28578352 | 4.18 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr21_+_33459524 | 4.11 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr1_+_16127825 | 4.03 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr8_+_23639124 | 4.01 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr20_-_20610812 | 3.93 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr14_-_32824380 | 3.92 |
ENSDART00000172791
ENSDART00000105745 |
inppl1b
|
inositol polyphosphate phosphatase-like 1b |
| chr14_+_29769336 | 3.86 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr13_+_27316934 | 3.84 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr5_-_25583125 | 3.83 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr10_+_39084354 | 3.80 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr13_+_27316632 | 3.79 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr17_-_15546862 | 3.76 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr9_-_24209083 | 3.76 |
ENSDART00000134599
|
zgc:153521
|
zgc:153521 |
| chr20_+_23238833 | 3.67 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr18_+_6857071 | 3.65 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr6_-_13022166 | 3.65 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr13_+_1100197 | 3.62 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr2_+_11205795 | 3.62 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr1_+_45969240 | 3.62 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr12_-_19346678 | 3.54 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr18_-_20560007 | 3.53 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr12_+_16967715 | 3.53 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr6_-_40744720 | 3.48 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr12_+_25775734 | 3.46 |
ENSDART00000024415
ENSDART00000149198 |
epas1a
|
endothelial PAS domain protein 1a |
| chr15_+_21252532 | 3.45 |
ENSDART00000162619
ENSDART00000019636 ENSDART00000144901 ENSDART00000138676 ENSDART00000133821 ENSDART00000146967 ENSDART00000143990 ENSDART00000142070 ENSDART00000132373 |
usf1
|
upstream transcription factor 1 |
| chr3_-_28258462 | 3.45 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr2_+_49572059 | 3.45 |
ENSDART00000108861
|
sema4e
|
semaphorin 4e |
| chr21_-_22435957 | 3.44 |
ENSDART00000137959
|
il7r
|
interleukin 7 receptor |
| chr5_+_34407763 | 3.39 |
ENSDART00000188849
ENSDART00000145127 |
lamc3
|
laminin, gamma 3 |
| chr25_-_11088839 | 3.33 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr21_+_45757317 | 3.29 |
ENSDART00000163152
|
h2afy
|
H2A histone family, member Y |
| chr25_+_19149241 | 3.28 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr2_-_11662851 | 3.27 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr5_-_51998708 | 3.26 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr15_+_26933196 | 3.20 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr19_-_34063567 | 3.19 |
ENSDART00000157815
ENSDART00000183907 |
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr11_-_30634286 | 3.17 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr18_-_1228688 | 3.15 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr3_+_15817644 | 3.11 |
ENSDART00000055787
|
natd1
|
zgc:110779 |
| chr12_-_28848015 | 3.11 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr5_-_66749535 | 3.10 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr4_+_18843015 | 3.08 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr14_+_26715626 | 3.08 |
ENSDART00000078557
|
polr2gl
|
polymerase (RNA) II (DNA directed) polypeptide G-like |
| chr5_-_46329880 | 3.03 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr11_+_16153207 | 3.02 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr10_+_26834985 | 2.99 |
ENSDART00000147518
|
fam89b
|
family with sequence similarity 89, member B |
| chr12_-_28537615 | 2.95 |
ENSDART00000067762
|
MYO1D
|
si:ch211-94l19.4 |
| chr16_+_10318893 | 2.94 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr7_+_22688781 | 2.91 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr20_-_40119872 | 2.90 |
ENSDART00000191569
|
nkain2
|
sodium/potassium transporting ATPase interacting 2 |
| chr12_+_8822717 | 2.89 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr22_+_30335936 | 2.84 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr19_+_46222428 | 2.83 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr5_+_22579975 | 2.82 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr2_-_12243213 | 2.79 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr14_-_32876280 | 2.78 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr20_-_19590378 | 2.78 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr1_-_30689004 | 2.77 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr17_+_44030692 | 2.77 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr9_+_17971935 | 2.76 |
ENSDART00000149736
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr3_-_31254379 | 2.76 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr1_-_37383741 | 2.75 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr20_-_31497300 | 2.72 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr22_-_3914162 | 2.69 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
| chr3_-_45298487 | 2.67 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr16_+_10413551 | 2.66 |
ENSDART00000032877
ENSDART00000172827 |
ino80e
|
INO80 complex subunit E |
| chr13_+_30951155 | 2.63 |
ENSDART00000057469
ENSDART00000162254 |
vstm4a
|
V-set and transmembrane domain containing 4a |
| chr11_-_36001495 | 2.63 |
ENSDART00000190330
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr11_+_16152316 | 2.60 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr14_-_48939560 | 2.58 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr6_-_8244474 | 2.56 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr5_+_37504309 | 2.56 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr5_-_45894802 | 2.55 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr17_+_38295847 | 2.54 |
ENSDART00000008532
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr3_+_10152092 | 2.51 |
ENSDART00000066053
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr18_+_41561285 | 2.50 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr11_-_16152400 | 2.49 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr17_+_24718272 | 2.48 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr14_+_26439227 | 2.48 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr19_+_46222918 | 2.48 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr3_+_31621774 | 2.47 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr24_-_25004553 | 2.47 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr25_-_25142387 | 2.47 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr3_-_25369557 | 2.46 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr23_+_31596441 | 2.45 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr12_-_33314894 | 2.43 |
ENSDART00000152908
|
c1qtnf1
|
C1q and TNF related 1 |
| chr14_+_33427837 | 2.43 |
ENSDART00000105687
|
zbtb33
|
zinc finger and BTB domain containing 33 |
| chr20_-_39367895 | 2.43 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr10_+_33754967 | 2.42 |
ENSDART00000153442
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr2_-_30200206 | 2.40 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr10_-_36633882 | 2.38 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr7_-_46782264 | 2.38 |
ENSDART00000166565
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr3_+_27798094 | 2.37 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr22_+_3914318 | 2.37 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr13_-_4333181 | 2.36 |
ENSDART00000122406
|
znf318
|
zinc finger protein 318 |
| chr3_-_26191960 | 2.35 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr11_+_11120532 | 2.34 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr7_+_56472585 | 2.33 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr19_-_32600823 | 2.33 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr4_+_9612574 | 2.32 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr16_+_6944564 | 2.32 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr20_+_25552057 | 2.31 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr15_+_37589698 | 2.30 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr14_-_33177935 | 2.30 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr18_+_16749091 | 2.28 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr17_+_15388479 | 2.24 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr5_-_56924747 | 2.23 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr6_-_59505589 | 2.23 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr3_+_19685873 | 2.23 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr24_-_6678640 | 2.19 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr19_+_10396042 | 2.18 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr3_+_52999962 | 2.18 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr3_-_60711127 | 2.17 |
ENSDART00000184119
|
ubald2
|
UBA-like domain containing 2 |
| chr23_-_10723009 | 2.17 |
ENSDART00000189721
|
foxp1a
|
forkhead box P1a |
| chr11_-_21586157 | 2.16 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr19_+_40115977 | 2.16 |
ENSDART00000139802
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr9_+_17348745 | 2.14 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr3_-_16039619 | 2.13 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr14_+_41406321 | 2.12 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr25_+_28823952 | 2.12 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr8_-_25716074 | 2.11 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr9_-_13355071 | 2.10 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr7_+_47287307 | 2.09 |
ENSDART00000114669
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr3_-_15496551 | 2.08 |
ENSDART00000124063
ENSDART00000007726 |
sgf29
|
SAGA complex associated factor 29 |
| chr3_-_15679107 | 2.04 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr11_+_6819050 | 2.03 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr9_+_41459759 | 2.02 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr11_-_34577034 | 2.01 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr21_-_42202792 | 2.01 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr18_+_1703984 | 2.00 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr11_-_25733910 | 1.99 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr13_+_9368621 | 1.98 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr19_+_20177887 | 1.95 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr17_-_50010121 | 1.95 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr17_+_24111392 | 1.95 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr4_-_14207471 | 1.93 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr6_-_32045951 | 1.92 |
ENSDART00000016629
ENSDART00000139055 |
efcab7
|
EF-hand calcium binding domain 7 |
| chr17_+_41992054 | 1.90 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr10_+_22890791 | 1.90 |
ENSDART00000176011
|
arrb2a
|
arrestin, beta 2a |
| chr6_-_53334259 | 1.90 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr13_-_22903246 | 1.89 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr23_-_17450746 | 1.89 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr1_+_25801648 | 1.87 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr6_-_40922971 | 1.86 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr6_-_42418225 | 1.86 |
ENSDART00000002501
|
ip6k2a
|
inositol hexakisphosphate kinase 2a |
| chr18_-_15911394 | 1.86 |
ENSDART00000091339
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr3_-_15496295 | 1.85 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr12_-_29624638 | 1.84 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr5_-_20446082 | 1.84 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr15_+_46313082 | 1.84 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr14_-_15155384 | 1.83 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr7_+_60359347 | 1.82 |
ENSDART00000145201
ENSDART00000039827 |
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr21_-_27273147 | 1.82 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr9_+_32358514 | 1.81 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr11_-_183328 | 1.80 |
ENSDART00000168562
|
avil
|
advillin |
| chr13_-_43599898 | 1.79 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr2_+_16696052 | 1.78 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr1_-_9277986 | 1.77 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.4 | 4.1 | GO:0010934 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 1.2 | 3.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.2 | 7.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.7 | 2.2 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.7 | 4.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.7 | 2.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.7 | 3.4 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.7 | 2.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.6 | 3.8 | GO:0002828 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) regulation of type 2 immune response(GO:0002828) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.6 | 5.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.6 | 7.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.6 | 2.8 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.5 | 3.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.5 | 1.9 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.5 | 1.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.5 | 1.4 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.5 | 1.9 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.5 | 2.8 | GO:0051818 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.5 | 6.8 | GO:0001964 | startle response(GO:0001964) |
| 0.4 | 1.3 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.4 | 2.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.4 | 2.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.4 | 2.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.4 | 2.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 1.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.4 | 2.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.4 | 1.2 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.4 | 1.9 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.4 | 1.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.4 | 1.5 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.4 | 2.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.4 | 2.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.3 | 12.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.3 | 2.6 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 4.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 2.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 4.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 8.8 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.3 | 1.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.3 | 0.8 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.3 | 2.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 2.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 1.3 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.3 | 1.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 1.5 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 3.5 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.2 | 3.5 | GO:0071456 | cellular response to decreased oxygen levels(GO:0036294) cellular response to hypoxia(GO:0071456) |
| 0.2 | 4.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.7 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.2 | 0.9 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.2 | 0.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 7.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 2.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 0.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 4.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 1.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 0.6 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 1.8 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.2 | 0.8 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.2 | 12.6 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.2 | 1.9 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.2 | 0.8 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.2 | 0.6 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 3.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.2 | 3.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.2 | 4.6 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.2 | 1.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.2 | 9.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.2 | 3.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 0.9 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 1.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 1.4 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 5.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 3.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 2.0 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 6.0 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.1 | 4.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.7 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 2.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 7.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 5.2 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 1.7 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 1.1 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.5 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 7.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 2.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.8 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.4 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 3.3 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.1 | 1.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 3.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.1 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.1 | 1.4 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.1 | 1.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.0 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 6.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 2.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 1.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.6 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 1.5 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 0.6 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 3.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 0.3 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 1.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 8.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 1.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 2.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.7 | GO:0045824 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.2 | GO:0036316 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 3.2 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 2.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 11.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 0.4 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.1 | 1.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 1.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 3.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.3 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 2.3 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.5 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 2.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.6 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 3.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.0 | GO:0010821 | regulation of mitochondrion organization(GO:0010821) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 2.1 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 1.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 3.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 1.0 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.4 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 1.8 | GO:0006906 | vesicle fusion(GO:0006906) organelle membrane fusion(GO:0090174) |
| 0.0 | 1.6 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 1.0 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 1.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 1.5 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 5.3 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 1.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.6 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.7 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 1.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.9 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 5.2 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
| 0.0 | 7.0 | GO:0031175 | neuron projection development(GO:0031175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 1.3 | 12.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.1 | 13.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.1 | 9.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.9 | 2.8 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.8 | 4.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 2.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.5 | 4.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 2.0 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.5 | 2.0 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.4 | 3.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 6.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 3.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 1.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 5.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 4.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 3.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 1.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 1.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 2.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 1.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 2.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 3.1 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 0.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 4.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 2.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.2 | 2.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 2.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 5.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 11.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 7.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 3.3 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 8.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 6.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 6.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 3.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.7 | GO:0001725 | stress fiber(GO:0001725) actomyosin(GO:0042641) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 5.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.9 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.9 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 7.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 7.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 26.9 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 3.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 3.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 3.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.2 | 4.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.8 | 2.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.8 | 4.9 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.6 | 3.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.6 | 1.9 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.6 | 3.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 3.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.5 | 9.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.5 | 4.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 1.4 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.4 | 1.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 6.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 1.6 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 6.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 10.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 1.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 3.5 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 1.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.3 | 3.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 1.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.3 | 2.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 0.9 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.3 | 2.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 7.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.3 | 1.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 4.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 3.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 1.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 11.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 1.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 1.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 4.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 4.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 6.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 0.9 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 1.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 3.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 7.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 3.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 2.0 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 8.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 7.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.9 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 3.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.9 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 3.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 1.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 3.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 2.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 1.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 5.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 2.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 2.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.9 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 2.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 4.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 6.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 3.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 3.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.0 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 6.4 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 7.3 | GO:0008528 | peptide receptor activity(GO:0001653) G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 2.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 3.3 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 3.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 5.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 9.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 3.6 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 1.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 3.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 12.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 3.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 5.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 6.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 3.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 1.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 6.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 3.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 7.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 2.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 2.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 12.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 2.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 4.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |