Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
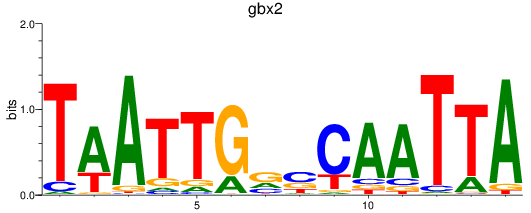
Results for gbx2
Z-value: 0.49
Transcription factors associated with gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gbx2
|
ENSDARG00000002933 | gastrulation brain homeobox 2 |
|
gbx2
|
ENSDARG00000116377 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gbx2 | dr11_v1_chr6_+_16031189_16031189 | 0.73 | 6.7e-17 | Click! |
Activity profile of gbx2 motif
Sorted Z-values of gbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_31821396 | 5.61 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr3_-_21037840 | 4.40 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr7_-_58098814 | 4.20 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr17_-_37214196 | 3.71 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr18_+_26895994 | 3.38 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr1_-_18803919 | 3.37 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr10_-_34741738 | 3.29 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr4_-_5019113 | 3.04 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr12_+_5977777 | 3.02 |
ENSDART00000152302
|
si:ch211-131k2.2
|
si:ch211-131k2.2 |
| chr17_+_24318753 | 2.85 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr13_+_733027 | 2.52 |
ENSDART00000149547
|
nrxn1b
|
neurexin 1b |
| chr1_+_40023640 | 2.49 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr11_+_6819050 | 2.45 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr4_-_5018705 | 2.31 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr18_+_16750080 | 2.30 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr8_+_21229718 | 2.25 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr10_+_43797130 | 2.15 |
ENSDART00000027242
|
nr2f1b
|
nuclear receptor subfamily 2, group F, member 1b |
| chr5_+_30624183 | 2.15 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr18_+_7639401 | 2.13 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr4_+_11384891 | 2.13 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr20_-_18382708 | 2.08 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr1_+_18811679 | 2.00 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr22_-_619711 | 1.96 |
ENSDART00000192778
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr21_-_43606502 | 1.91 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr15_-_10341048 | 1.89 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr5_+_57773222 | 1.83 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr22_+_24157807 | 1.79 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr9_-_9671244 | 1.79 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr7_-_33684632 | 1.74 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr18_+_17537344 | 1.74 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr16_+_5678071 | 1.72 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr11_-_2632747 | 1.72 |
ENSDART00000008133
|
si:ch211-160o17.2
|
si:ch211-160o17.2 |
| chr18_+_31057143 | 1.71 |
ENSDART00000164585
|
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr18_-_16937008 | 1.68 |
ENSDART00000100117
ENSDART00000022640 ENSDART00000136541 |
znf143b
|
zinc finger protein 143b |
| chr11_+_24620742 | 1.68 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr16_-_36093776 | 1.66 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr12_-_35386910 | 1.60 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr2_-_23004286 | 1.60 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr8_+_23105117 | 1.59 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr7_+_59677273 | 1.44 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
| chr17_+_10593398 | 1.43 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr1_-_18811517 | 1.39 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr19_-_20430892 | 1.36 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr22_-_36530902 | 1.31 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_-_22512063 | 1.30 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr23_-_333457 | 1.28 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr14_+_35464994 | 1.25 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr17_+_49281597 | 1.23 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr23_+_4253957 | 1.21 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr15_-_16177603 | 1.18 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr5_-_20123002 | 1.14 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr8_+_20140321 | 1.12 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr22_+_2417105 | 1.11 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr5_-_69923241 | 1.09 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr8_+_23826985 | 1.08 |
ENSDART00000187430
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr10_-_32920690 | 1.06 |
ENSDART00000136245
|
cux1a
|
cut-like homeobox 1a |
| chr11_-_12158412 | 1.00 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr14_+_25816874 | 0.97 |
ENSDART00000005499
|
glra1
|
glycine receptor, alpha 1 |
| chr6_+_3717613 | 0.95 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr24_-_25364775 | 0.95 |
ENSDART00000151915
|
ptchd1
|
patched domain containing 1 |
| chr16_-_26296477 | 0.89 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr15_-_11956981 | 0.88 |
ENSDART00000164163
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr11_+_24340166 | 0.88 |
ENSDART00000188719
|
rbm39a
|
RNA binding motif protein 39a |
| chr3_+_32118670 | 0.88 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr10_-_14929630 | 0.86 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr8_+_18010568 | 0.85 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr23_+_42254960 | 0.81 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr8_-_18010097 | 0.81 |
ENSDART00000122730
ENSDART00000133666 |
acot11b
|
acyl-CoA thioesterase 11b |
| chr24_+_33802528 | 0.74 |
ENSDART00000136040
ENSDART00000147499 ENSDART00000182322 |
atg9b
|
autophagy related 9B |
| chr16_+_54875530 | 0.67 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr23_-_30727596 | 0.64 |
ENSDART00000060193
|
thap3
|
THAP domain containing, apoptosis associated protein 3 |
| chr2_+_35240485 | 0.62 |
ENSDART00000179804
|
tnr
|
tenascin R (restrictin, janusin) |
| chr11_-_422484 | 0.58 |
ENSDART00000122253
|
znf831
|
zinc finger protein 831 |
| chr17_+_51499789 | 0.57 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr19_-_15855427 | 0.55 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr17_+_1496107 | 0.54 |
ENSDART00000187804
|
LO018430.1
|
|
| chr22_+_9862466 | 0.53 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr22_+_9862243 | 0.49 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr20_-_7128612 | 0.47 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr6_-_42369186 | 0.34 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr11_+_31323746 | 0.33 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr6_-_39218609 | 0.33 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr11_-_6452444 | 0.32 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr7_+_53755054 | 0.28 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr22_-_9861531 | 0.27 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr18_+_8833251 | 0.26 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr12_-_34713690 | 0.26 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr25_-_20523927 | 0.25 |
ENSDART00000114448
|
si:ch211-269c21.2
|
si:ch211-269c21.2 |
| chr24_-_33801668 | 0.25 |
ENSDART00000079202
|
abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr2_-_26720854 | 0.25 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr24_-_16904234 | 0.22 |
ENSDART00000192879
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr7_+_20260172 | 0.21 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr8_+_8298439 | 0.17 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr17_+_41302660 | 0.16 |
ENSDART00000059480
|
fosl2
|
fos-like antigen 2 |
| chr6_+_9191981 | 0.10 |
ENSDART00000150916
|
zgc:112392
|
zgc:112392 |
| chr9_+_55154414 | 0.05 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr15_+_28355023 | 0.05 |
ENSDART00000122159
|
si:dkey-118k5.3
|
si:dkey-118k5.3 |
| chr2_-_36918709 | 0.02 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr11_-_29768054 | 0.01 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr9_-_30555725 | 0.01 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr12_+_22560067 | 0.00 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
Network of associatons between targets according to the STRING database.
First level regulatory network of gbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.8 | 2.4 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.7 | 4.4 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.6 | 3.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.5 | 5.3 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.4 | 1.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 1.9 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 1.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.3 | 1.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.2 | 1.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.2 | 2.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 2.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 1.1 | GO:0040016 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.6 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 2.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.8 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.9 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 3.4 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 2.2 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 2.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.2 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 1.7 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 3.4 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.9 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 5.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.1 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.2 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.2 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 3.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 2.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 3.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.0 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.5 | 3.4 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.5 | 1.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.7 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.3 | 3.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.3 | 1.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 4.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 1.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 2.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.2 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.9 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 5.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 3.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.4 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 2.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |