Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
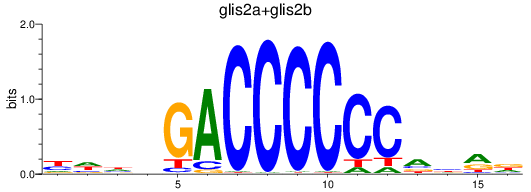
Results for glis2a+glis2b
Z-value: 0.48
Transcription factors associated with glis2a+glis2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
glis2a
|
ENSDARG00000078388 | GLIS family zinc finger 2a |
|
glis2b
|
ENSDARG00000100232 | GLIS family zinc finger 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| glis2b | dr11_v1_chr3_+_12334509_12334509 | -0.20 | 4.9e-02 | Click! |
| glis2a | dr11_v1_chr22_-_26558166_26558166 | 0.11 | 3.1e-01 | Click! |
Activity profile of glis2a+glis2b motif
Sorted Z-values of glis2a+glis2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_4070058 | 5.75 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr24_+_25471196 | 5.20 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr23_-_17470146 | 2.84 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr23_-_45705525 | 2.75 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr23_+_42810055 | 2.05 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr7_+_18364176 | 2.02 |
ENSDART00000171606
ENSDART00000186368 |
cd248a
|
CD248 molecule, endosialin a |
| chr2_-_30987538 | 1.99 |
ENSDART00000148157
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr9_+_20781047 | 1.86 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr17_-_20717845 | 1.85 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr25_+_13191615 | 1.61 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr19_-_15281996 | 1.61 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr20_+_25340814 | 1.60 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr5_-_42272517 | 1.60 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr23_-_29004753 | 1.47 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr25_+_13191391 | 1.44 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr5_+_51079504 | 1.40 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr8_+_7033049 | 1.36 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr10_+_33982010 | 1.32 |
ENSDART00000180431
|
fryb
|
furry homolog b (Drosophila) |
| chr10_-_34976812 | 1.23 |
ENSDART00000170625
|
smad9
|
SMAD family member 9 |
| chr7_+_38509333 | 1.12 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr10_-_13116337 | 0.98 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr10_-_1961576 | 0.98 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr23_-_29003864 | 0.95 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr5_-_71995108 | 0.79 |
ENSDART00000124587
|
fam78ab
|
family with sequence similarity 78, member Ab |
| chr19_-_42928979 | 0.78 |
ENSDART00000086659
|
cyhr1
|
cysteine/histidine-rich 1 |
| chr12_+_22672323 | 0.78 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr23_+_43668756 | 0.73 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr17_-_4245902 | 0.65 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr6_+_37625787 | 0.61 |
ENSDART00000065122
|
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr3_-_31804481 | 0.60 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr16_+_3004422 | 0.51 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr18_+_33100606 | 0.49 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr19_+_15443540 | 0.48 |
ENSDART00000193355
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr23_-_31060350 | 0.48 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr16_-_41059329 | 0.45 |
ENSDART00000145556
|
si:dkey-201i6.8
|
si:dkey-201i6.8 |
| chr24_-_38574631 | 0.43 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr7_-_24838857 | 0.41 |
ENSDART00000179766
ENSDART00000180892 |
fam113
|
family with sequence similarity 113 |
| chr6_+_38845848 | 0.41 |
ENSDART00000184907
|
stk35l
|
serine/threonine kinase 35, like |
| chr19_+_20540843 | 0.41 |
ENSDART00000192500
|
kcnh8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr17_-_4245311 | 0.40 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr2_-_9744081 | 0.40 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr6_+_38845697 | 0.39 |
ENSDART00000053187
|
stk35l
|
serine/threonine kinase 35, like |
| chr20_+_7799615 | 0.37 |
ENSDART00000152792
|
si:dkey-19f4.2
|
si:dkey-19f4.2 |
| chr17_-_49275690 | 0.37 |
ENSDART00000150172
|
tfb1m
|
transcription factor B1, mitochondrial |
| chr13_-_24880525 | 0.36 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr19_-_36640677 | 0.36 |
ENSDART00000189743
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr18_+_17827149 | 0.35 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr24_-_9960290 | 0.33 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr5_-_1047504 | 0.32 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr5_+_38886499 | 0.32 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr12_-_145076 | 0.31 |
ENSDART00000160926
|
dnah9
|
dynein, axonemal, heavy chain 9 |
| chr12_+_2676878 | 0.30 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr22_-_18116635 | 0.29 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr3_+_8826905 | 0.28 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr13_-_638485 | 0.24 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr20_-_1239596 | 0.22 |
ENSDART00000049063
ENSDART00000140650 |
ankrd6b
|
ankyrin repeat domain 6b |
| chr4_-_4756349 | 0.21 |
ENSDART00000057507
|
ssbp1
|
single-stranded DNA binding protein 1 |
| chr8_-_53926228 | 0.21 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr23_-_26536055 | 0.21 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr7_+_11390462 | 0.20 |
ENSDART00000114383
|
tlnrd1
|
talin rod domain containing 1 |
| chr8_-_20914829 | 0.20 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr13_+_1928361 | 0.19 |
ENSDART00000164764
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr12_-_41619257 | 0.18 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr25_+_30196039 | 0.18 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr5_-_22001003 | 0.15 |
ENSDART00000134393
ENSDART00000143878 |
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr22_+_26853254 | 0.15 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr7_-_21928826 | 0.14 |
ENSDART00000088043
|
si:dkey-85k7.11
|
si:dkey-85k7.11 |
| chr15_-_97205 | 0.14 |
ENSDART00000158734
|
BACE1
|
beta-secretase 1 |
| chr25_-_997894 | 0.13 |
ENSDART00000169011
|
znf609a
|
zinc finger protein 609a |
| chr18_+_17151916 | 0.10 |
ENSDART00000175488
|
trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr2_+_51783120 | 0.09 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr10_+_2742499 | 0.05 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr16_-_45854882 | 0.03 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr4_-_1776352 | 0.02 |
ENSDART00000123089
|
dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr15_+_518092 | 0.00 |
ENSDART00000154594
|
pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of glis2a+glis2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.4 | 2.8 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.3 | 1.4 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 1.0 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 5.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 0.7 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 1.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.4 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.6 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.2 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 0.6 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 3.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 2.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 1.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 2.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.4 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 1.0 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 1.1 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.6 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |