Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
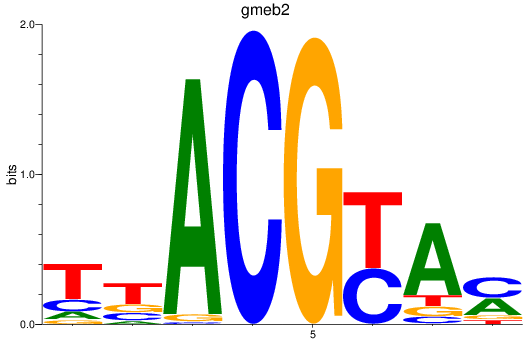
Results for gmeb2
Z-value: 1.02
Transcription factors associated with gmeb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gmeb2
|
ENSDARG00000093240 | si_ch73-302a13.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | dr11_v1_chr23_-_41821825_41821825 | 0.73 | 3.4e-17 | Click! |
Activity profile of gmeb2 motif
Sorted Z-values of gmeb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_46111849 | 16.57 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr20_-_27381691 | 11.11 |
ENSDART00000010293
|
serpina10a
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10a |
| chr2_-_54387550 | 9.45 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr8_-_4618653 | 8.46 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr16_+_28596555 | 8.46 |
ENSDART00000046209
ENSDART00000141708 |
acbd7
|
acyl-CoA binding domain containing 7 |
| chr1_-_56223913 | 7.94 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr15_-_12319065 | 7.29 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr11_+_37275448 | 7.21 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr13_-_21688176 | 7.19 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr1_+_25801648 | 7.05 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr17_+_27176243 | 6.99 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr19_+_46259619 | 6.75 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr4_+_45148652 | 6.72 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr3_-_29910547 | 6.61 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr3_-_13147310 | 6.48 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr11_-_36475124 | 6.35 |
ENSDART00000165203
|
usp48
|
ubiquitin specific peptidase 48 |
| chr6_-_11780070 | 6.33 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr11_-_41132296 | 6.25 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr13_+_4405282 | 6.23 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr5_+_24245682 | 6.17 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr1_-_8101495 | 5.94 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr6_-_40744720 | 5.83 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr19_-_31402429 | 5.81 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr18_-_40481028 | 5.77 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr5_+_3501859 | 5.77 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr24_+_5208171 | 5.72 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr22_-_11493236 | 5.68 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr14_-_43000836 | 5.64 |
ENSDART00000162714
|
pcdh10b
|
protocadherin 10b |
| chr25_-_29988352 | 5.50 |
ENSDART00000067059
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr23_+_17899226 | 5.42 |
ENSDART00000079487
|
CU234171.1
|
|
| chr20_-_45661049 | 5.35 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr8_+_31941650 | 5.34 |
ENSDART00000138217
|
htr1aa
|
5-hydroxytryptamine (serotonin) receptor 1A a |
| chr3_-_13146631 | 5.16 |
ENSDART00000172460
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr20_+_25486206 | 5.10 |
ENSDART00000172076
|
hook1
|
hook microtubule-tethering protein 1 |
| chr2_-_21170517 | 5.05 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr24_+_20968308 | 4.79 |
ENSDART00000130568
|
drd3
|
dopamine receptor D3 |
| chr8_-_25120231 | 4.72 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr3_+_32789605 | 4.67 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr3_-_46410387 | 4.64 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr20_+_34717403 | 4.43 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr3_-_25814097 | 4.19 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr17_-_51818659 | 4.15 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr1_-_6494384 | 4.14 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr20_-_44576949 | 4.12 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr13_+_12299997 | 4.06 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr8_+_53311965 | 4.06 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr21_-_43040780 | 4.00 |
ENSDART00000187037
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr18_+_11970987 | 3.92 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr18_+_31410652 | 3.90 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr20_+_25486021 | 3.89 |
ENSDART00000063052
|
hook1
|
hook microtubule-tethering protein 1 |
| chr2_+_55914699 | 3.83 |
ENSDART00000157905
|
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr19_-_10395683 | 3.81 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr14_-_2358774 | 3.76 |
ENSDART00000164809
|
si:ch73-233f7.5
|
si:ch73-233f7.5 |
| chr7_+_13382852 | 3.74 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr9_+_24920677 | 3.72 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_-_10192459 | 3.69 |
ENSDART00000128535
ENSDART00000017173 |
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr17_-_31611692 | 3.60 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr16_-_20294236 | 3.56 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr21_-_33995710 | 3.54 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr3_+_32933663 | 3.43 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr2_-_31614277 | 3.29 |
ENSDART00000137054
ENSDART00000131507 ENSDART00000137674 ENSDART00000147041 |
si:ch211-106h4.5
|
si:ch211-106h4.5 |
| chr7_-_39751540 | 3.24 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr25_-_32464149 | 3.10 |
ENSDART00000152787
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr7_-_55539738 | 3.08 |
ENSDART00000168721
ENSDART00000013796 ENSDART00000148514 |
aprt
|
adenine phosphoribosyltransferase |
| chr11_-_18799827 | 3.05 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr11_+_29770966 | 2.97 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr6_-_37469775 | 2.95 |
ENSDART00000156546
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr5_+_26138313 | 2.92 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr17_+_16873417 | 2.91 |
ENSDART00000146276
|
dio2
|
iodothyronine deiodinase 2 |
| chr13_+_3938526 | 2.78 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr4_+_77053467 | 2.75 |
ENSDART00000187643
ENSDART00000150848 |
znf1009
|
zinc finger protein 1009 |
| chr8_-_19467011 | 2.73 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr16_+_40131473 | 2.68 |
ENSDART00000155421
ENSDART00000134732 ENSDART00000138699 |
cenpw
si:ch211-195p4.4
|
centromere protein W si:ch211-195p4.4 |
| chr15_+_11683114 | 2.67 |
ENSDART00000168233
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr12_+_17603528 | 2.62 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr15_-_11683529 | 2.62 |
ENSDART00000161445
|
fkrp
|
fukutin related protein |
| chr2_-_32826108 | 2.61 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr6_-_17849786 | 2.61 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr25_-_28668776 | 2.61 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr7_+_51795667 | 2.59 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr15_+_32798333 | 2.58 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr2_-_32486080 | 2.58 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr8_+_20455134 | 2.50 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr11_-_22372072 | 2.46 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr12_+_19401854 | 2.45 |
ENSDART00000153415
|
si:dkey-16i5.8
|
si:dkey-16i5.8 |
| chr15_+_5116179 | 2.45 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr9_+_21819082 | 2.43 |
ENSDART00000136902
ENSDART00000101991 |
txndc9
|
thioredoxin domain containing 9 |
| chr20_-_30938184 | 2.42 |
ENSDART00000147359
ENSDART00000062552 |
wtap
|
WT1 associated protein |
| chr23_-_38330746 | 2.41 |
ENSDART00000192262
|
FO904868.1
|
|
| chr24_-_7826489 | 2.40 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr9_-_23242684 | 2.34 |
ENSDART00000053282
ENSDART00000179770 |
ccnt2a
|
cyclin T2a |
| chr2_-_3403020 | 2.33 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr14_-_24095414 | 2.30 |
ENSDART00000172747
ENSDART00000173146 |
si:ch211-277c7.7
|
si:ch211-277c7.7 |
| chr14_+_8947282 | 2.28 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr21_-_33995213 | 2.24 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr3_+_46764278 | 2.21 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr5_-_23596339 | 2.20 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr10_-_41907213 | 2.19 |
ENSDART00000167004
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr6_-_51541488 | 2.18 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr16_+_41015163 | 2.16 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr15_-_20412286 | 2.16 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr9_-_4598883 | 2.07 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr9_+_19529951 | 2.05 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr19_+_40026041 | 2.04 |
ENSDART00000017359
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr12_+_11650146 | 1.99 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr5_-_54395488 | 1.99 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr8_-_30944465 | 1.97 |
ENSDART00000128792
ENSDART00000191717 ENSDART00000049944 |
smarcb1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1a |
| chr20_+_38837238 | 1.97 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr10_-_14545658 | 1.96 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr25_-_36044583 | 1.96 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr8_+_47897734 | 1.94 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr18_-_46208581 | 1.93 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr11_+_30000814 | 1.90 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr9_+_12948511 | 1.87 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr11_+_2506516 | 1.86 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr20_-_32405440 | 1.78 |
ENSDART00000062978
ENSDART00000153411 |
afg1lb
|
AFG1 like ATPase b |
| chr20_-_9199721 | 1.73 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr3_+_46763745 | 1.73 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr1_+_26105141 | 1.71 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr3_+_46764022 | 1.67 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr4_+_23126558 | 1.67 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr6_+_9163007 | 1.66 |
ENSDART00000183054
ENSDART00000157552 |
zgc:112023
|
zgc:112023 |
| chr2_+_2168547 | 1.63 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr25_-_8513255 | 1.63 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr8_+_13064750 | 1.61 |
ENSDART00000039878
|
sap30bp
|
SAP30 binding protein |
| chr10_+_43088270 | 1.60 |
ENSDART00000191221
|
xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr1_+_23274710 | 1.56 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr12_-_27588299 | 1.55 |
ENSDART00000178023
ENSDART00000066282 |
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr16_-_47427016 | 1.53 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr1_+_49415281 | 1.44 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr7_+_4162994 | 1.43 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr17_-_43399896 | 1.38 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr14_-_30141162 | 1.37 |
ENSDART00000053916
|
mtnr1ab
|
melatonin receptor 1A b |
| chr9_+_9502610 | 1.36 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr19_-_35155722 | 1.35 |
ENSDART00000151924
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr9_-_28103097 | 1.34 |
ENSDART00000146284
|
kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr6_-_39903393 | 1.33 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr7_+_57341805 | 1.32 |
ENSDART00000115222
|
rxfp3.3a2
|
relaxin/insulin-like family peptide receptor 3.3a2 |
| chr5_-_51198430 | 1.32 |
ENSDART00000132503
ENSDART00000097473 ENSDART00000165870 |
snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr11_-_14813029 | 1.30 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr14_+_6605675 | 1.29 |
ENSDART00000143179
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr14_-_21932403 | 1.26 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr17_-_25831569 | 1.26 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr7_+_55292959 | 1.23 |
ENSDART00000147539
ENSDART00000073555 |
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr14_-_22495604 | 1.21 |
ENSDART00000137167
|
si:ch211-107m4.1
|
si:ch211-107m4.1 |
| chr11_+_11120532 | 1.18 |
ENSDART00000026135
ENSDART00000189872 |
ly75
|
lymphocyte antigen 75 |
| chr5_-_16139600 | 1.18 |
ENSDART00000051644
|
coq5
|
coenzyme Q5, methyltransferase |
| chr6_-_40455109 | 1.12 |
ENSDART00000103878
|
vhl
|
von Hippel-Lindau tumor suppressor |
| chr7_+_41313568 | 1.11 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr2_+_52049239 | 1.09 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr13_-_12486076 | 1.07 |
ENSDART00000146195
|
lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr20_-_20312789 | 1.04 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr23_+_2703044 | 1.03 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr9_+_35860975 | 1.00 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr25_+_21098675 | 0.99 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr10_+_29137482 | 0.98 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr19_+_19241372 | 0.97 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr5_-_13076779 | 0.95 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr2_+_32826235 | 0.94 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr16_-_47426482 | 0.94 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr25_-_21066136 | 0.92 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr7_+_66634167 | 0.87 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr8_-_5267442 | 0.84 |
ENSDART00000155816
|
ppp2r2aa
|
protein phosphatase 2, regulatory subunit B, alpha a |
| chr20_+_2227860 | 0.83 |
ENSDART00000152321
|
tmem200a
|
transmembrane protein 200A |
| chr22_-_621609 | 0.79 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr13_+_21676235 | 0.76 |
ENSDART00000137804
ENSDART00000134950 ENSDART00000129653 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr7_-_7493758 | 0.75 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr16_-_1502699 | 0.75 |
ENSDART00000187189
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr23_-_35790235 | 0.73 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr12_+_28956374 | 0.70 |
ENSDART00000033878
|
znf668
|
zinc finger protein 668 |
| chr1_+_9860381 | 0.69 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr5_-_24245218 | 0.64 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr2_+_10821127 | 0.61 |
ENSDART00000145770
ENSDART00000174629 ENSDART00000081094 |
glmna
|
glomulin, FKBP associated protein a |
| chr4_-_11810799 | 0.60 |
ENSDART00000014153
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr19_-_432083 | 0.60 |
ENSDART00000165371
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr25_+_21098990 | 0.59 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr25_-_17494364 | 0.57 |
ENSDART00000154134
|
lrrc29
|
leucine rich repeat containing 29 |
| chr22_-_621888 | 0.56 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr13_+_36923052 | 0.56 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr23_+_20640484 | 0.54 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr13_-_36535128 | 0.54 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr10_-_15910974 | 0.54 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr3_-_30186296 | 0.52 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr5_+_57210237 | 0.52 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr22_+_26798853 | 0.49 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr3_-_8765165 | 0.49 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr12_+_33460794 | 0.46 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr3_+_26342768 | 0.45 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr17_-_37195163 | 0.45 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr20_+_25563105 | 0.45 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr4_-_11810365 | 0.43 |
ENSDART00000150529
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr7_-_38570878 | 0.41 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr2_+_10821579 | 0.36 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr21_-_37733287 | 0.36 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr17_-_6076084 | 0.33 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr10_+_16501699 | 0.32 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr22_+_20145036 | 0.31 |
ENSDART00000137624
|
eef2a.2
|
eukaryotic translation elongation factor 2a, tandem duplicate 2 |
| chr9_+_25840720 | 0.31 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr17_-_12058171 | 0.29 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr8_+_23703680 | 0.29 |
ENSDART00000141099
ENSDART00000135394 |
ppardb
|
peroxisome proliferator-activated receptor delta b |
Network of associatons between targets according to the STRING database.
First level regulatory network of gmeb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.4 | 4.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 1.3 | 5.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 1.2 | 3.7 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.9 | 3.7 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.9 | 2.7 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.8 | 3.1 | GO:0006168 | adenine salvage(GO:0006168) |
| 0.8 | 12.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.7 | 2.9 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.7 | 2.0 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.6 | 5.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.6 | 6.7 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.6 | 9.0 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.5 | 1.6 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.5 | 2.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.5 | 1.6 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.5 | 3.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.5 | 1.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.5 | 2.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.4 | 1.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.4 | 1.3 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.4 | 2.6 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.4 | 2.9 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.4 | 4.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.4 | 2.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.4 | 1.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.4 | 5.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 3.8 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 4.8 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 | 5.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.3 | 1.7 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.3 | 1.6 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.3 | 6.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.3 | 5.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.3 | 2.0 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.3 | 1.9 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.3 | 1.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.3 | 2.0 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 | 0.7 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.2 | 3.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 5.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 5.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) serotonin receptor signaling pathway(GO:0007210) |
| 0.2 | 4.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 1.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 1.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 3.7 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.2 | 1.2 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 3.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 4.7 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 1.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.2 | 1.6 | GO:0032042 | mitochondrial DNA replication(GO:0006264) mitochondrial DNA metabolic process(GO:0032042) |
| 0.2 | 5.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 2.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 7.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.4 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 3.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.5 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 18.9 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.2 | GO:0045911 | positive regulation of DNA recombination(GO:0045911) |
| 0.1 | 1.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 10.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 3.7 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 9.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 2.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.7 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 3.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.7 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 1.0 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 4.4 | GO:0071696 | ectodermal placode development(GO:0071696) |
| 0.1 | 2.2 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.1 | 1.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.8 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 2.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 4.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 6.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 5.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 3.4 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.8 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 2.5 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 1.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.0 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 6.7 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 9.0 | GO:0070695 | FHF complex(GO:0070695) |
| 1.4 | 5.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 1.3 | 5.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.3 | 11.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.8 | 3.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.6 | 2.4 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.6 | 6.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 1.6 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.4 | 1.3 | GO:0072380 | TRC complex(GO:0072380) |
| 0.4 | 6.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 2.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 1.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 10.9 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.3 | 2.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.3 | 2.3 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 2.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 5.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 16.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 1.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 1.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 2.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 9.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 4.4 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 7.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 3.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.9 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 4.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 7.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 3.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 5.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.2 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 1.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.8 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 4.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 5.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 4.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 7.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 21.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 14.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.7 | 11.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.0 | 3.1 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 1.0 | 5.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.9 | 2.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.7 | 3.6 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.6 | 6.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 5.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.5 | 1.4 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.4 | 5.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.4 | 7.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 3.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.3 | 4.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.3 | 4.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.3 | 3.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 3.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 1.0 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.3 | 8.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.3 | 1.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.3 | 4.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 7.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 3.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.3 | 1.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 5.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 9.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 3.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 2.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 5.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 2.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 2.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 7.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 4.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 6.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 15.9 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 2.5 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 7.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 2.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 6.4 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 3.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 5.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 2.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 1.3 | GO:0030551 | cyclic nucleotide binding(GO:0030551) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 1.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.3 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 4.0 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 2.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 5.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.6 | 5.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.4 | 2.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.4 | 4.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 3.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 4.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 1.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.3 | 3.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 2.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 3.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 1.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |