Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
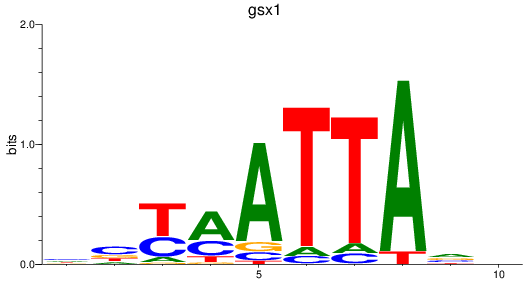
Results for gsx1
Z-value: 1.16
Transcription factors associated with gsx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsx1
|
ENSDARG00000035735 | GS homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsx1 | dr11_v1_chr5_-_67911111_67911111 | -0.80 | 6.9e-22 | Click! |
Activity profile of gsx1 motif
Sorted Z-values of gsx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_28445052 | 26.34 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr25_+_31276842 | 18.41 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr21_-_23110841 | 14.87 |
ENSDART00000147896
ENSDART00000003076 ENSDART00000184925 ENSDART00000190386 |
usp28
|
ubiquitin specific peptidase 28 |
| chr12_+_3078221 | 14.77 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr7_+_20503344 | 11.18 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr15_+_46357080 | 11.03 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr9_-_6372535 | 10.36 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr22_-_10121880 | 9.91 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr24_-_40744672 | 9.64 |
ENSDART00000160672
|
CU633479.1
|
|
| chr6_-_46941245 | 9.15 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr6_+_41191482 | 8.90 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr2_-_23768818 | 8.42 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr5_-_33287691 | 8.42 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr6_-_3982783 | 8.33 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr22_-_10586191 | 8.19 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr15_+_46356879 | 8.16 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr7_+_17816006 | 8.15 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr5_+_27897504 | 8.03 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr8_-_23612462 | 8.02 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr15_-_23376541 | 7.83 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr8_+_23738122 | 7.65 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr8_-_18537866 | 7.58 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr2_-_51511494 | 7.43 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr17_+_8799451 | 7.43 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr9_-_21918963 | 7.40 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr13_+_29238850 | 7.19 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr24_-_37640705 | 7.11 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr2_+_33326522 | 7.10 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr6_-_46861676 | 7.01 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr24_-_2450597 | 6.76 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr11_+_24703108 | 6.58 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr7_+_20344222 | 6.49 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr17_+_16046132 | 6.32 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr6_-_40581376 | 6.16 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr19_+_43297546 | 6.07 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr17_+_16046314 | 5.49 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr21_-_7035599 | 5.40 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr24_+_19415124 | 5.37 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr2_+_20793982 | 5.32 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr8_+_11425048 | 5.30 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr22_+_19366866 | 5.18 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr23_+_44374041 | 5.14 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr18_-_6982499 | 5.07 |
ENSDART00000101525
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr21_-_40174647 | 5.06 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr10_-_21362320 | 5.01 |
ENSDART00000189789
|
avd
|
avidin |
| chr12_+_20352400 | 4.99 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr2_-_23778180 | 4.93 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr16_-_24605969 | 4.79 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr15_-_37104165 | 4.59 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr7_+_17816470 | 4.55 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr7_+_2236317 | 4.54 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr12_+_10631266 | 4.51 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr14_+_22022441 | 4.49 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr10_+_16584382 | 4.49 |
ENSDART00000112039
|
CR790388.1
|
|
| chr16_-_28878080 | 4.47 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr13_+_35637048 | 4.39 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr15_-_43284021 | 4.38 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr6_-_39024538 | 4.37 |
ENSDART00000165839
|
tns2b
|
tensin 2b |
| chr4_-_51460013 | 4.34 |
ENSDART00000193382
|
CR628395.1
|
|
| chr25_+_32755485 | 4.22 |
ENSDART00000162188
|
etfa
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr22_+_20427170 | 4.18 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr12_+_32199651 | 4.13 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr5_-_16472719 | 4.12 |
ENSDART00000162071
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr15_-_41457898 | 4.12 |
ENSDART00000188847
|
nlrc9
|
NLR family CARD domain containing 9 |
| chr20_+_38458084 | 4.07 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr3_+_32112004 | 4.01 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr14_-_21959712 | 3.97 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr10_-_21362071 | 3.87 |
ENSDART00000125167
|
avd
|
avidin |
| chr8_+_21353878 | 3.85 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr18_+_2228737 | 3.82 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr14_+_33329761 | 3.79 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr8_+_54274355 | 3.75 |
ENSDART00000067639
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr15_-_25518084 | 3.68 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr15_-_19705707 | 3.67 |
ENSDART00000047643
|
sytl2b
|
synaptotagmin-like 2b |
| chr8_-_25336589 | 3.64 |
ENSDART00000009682
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr19_-_82504 | 3.57 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr20_-_40755614 | 3.56 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr10_+_40633990 | 3.51 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr11_-_2478374 | 3.49 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr1_-_58505626 | 3.47 |
ENSDART00000171304
|
si:ch73-236c18.8
|
si:ch73-236c18.8 |
| chr12_-_18577983 | 3.47 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr15_-_1822548 | 3.40 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr12_+_48803098 | 3.28 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr5_-_44286987 | 3.27 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr11_+_45323455 | 3.26 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr3_+_32129632 | 3.24 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr12_-_33770299 | 3.24 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr20_+_10723292 | 3.19 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr9_+_41156818 | 3.18 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr15_+_5293785 | 3.11 |
ENSDART00000163157
|
or121-1
|
odorant receptor, family E, subfamily 121, member 1 |
| chr24_+_6107901 | 3.04 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr16_-_17347727 | 3.03 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr11_+_36665359 | 2.96 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr4_-_42408339 | 2.91 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr4_+_43246676 | 2.89 |
ENSDART00000187588
|
si:dkey-29j8.1
|
si:dkey-29j8.1 |
| chr20_-_52902693 | 2.89 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr7_-_7764287 | 2.88 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr10_+_31646020 | 2.86 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr14_+_7377552 | 2.84 |
ENSDART00000142158
ENSDART00000141471 |
hars
|
histidyl-tRNA synthetase |
| chr19_+_10339538 | 2.84 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr4_+_52356485 | 2.81 |
ENSDART00000170639
|
zgc:173705
|
zgc:173705 |
| chr1_+_47178529 | 2.77 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr21_-_22827548 | 2.75 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr24_-_6078222 | 2.73 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr10_+_40756352 | 2.67 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr20_-_23426339 | 2.62 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr6_+_52891947 | 2.61 |
ENSDART00000174159
|
BX649282.3
|
|
| chr21_+_42708621 | 2.60 |
ENSDART00000158392
|
si:ch1073-204b8.1
|
si:ch1073-204b8.1 |
| chr11_-_45138857 | 2.60 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr8_+_37527575 | 2.58 |
ENSDART00000147239
|
or135-1
|
odorant receptor, family H, subfamily 135, member 1 |
| chr8_+_25034544 | 2.57 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr1_+_55239160 | 2.57 |
ENSDART00000152318
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr3_-_29941357 | 2.52 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr17_+_53425037 | 2.52 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr15_+_34988148 | 2.52 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr4_-_71210030 | 2.51 |
ENSDART00000186473
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr8_-_52229462 | 2.50 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr17_+_2162916 | 2.46 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr1_-_9123465 | 2.46 |
ENSDART00000081337
|
ndufab1a
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a |
| chr4_+_57194439 | 2.46 |
ENSDART00000075265
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr21_-_32060993 | 2.45 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr17_-_48944465 | 2.44 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr16_-_16761164 | 2.42 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr7_+_1467863 | 2.41 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr12_+_16168342 | 2.41 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr15_-_4969525 | 2.39 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr10_+_40660772 | 2.34 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr18_+_619619 | 2.29 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr11_+_14284866 | 2.29 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr25_+_5068442 | 2.22 |
ENSDART00000097522
|
parvg
|
parvin, gamma |
| chr20_-_9095105 | 2.21 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr20_+_25879826 | 2.19 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr3_-_32169754 | 2.19 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr23_+_11285662 | 2.18 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr7_+_48667081 | 2.16 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr23_-_41965557 | 2.15 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr2_-_38256589 | 2.12 |
ENSDART00000173064
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr21_-_17296789 | 2.12 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr3_+_32553714 | 2.10 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr10_+_2799285 | 2.09 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr15_+_1796313 | 2.06 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr8_-_17167819 | 2.01 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr17_+_46818521 | 1.98 |
ENSDART00000156022
|
pimr14
|
Pim proto-oncogene, serine/threonine kinase, related 14 |
| chr2_-_22927958 | 1.96 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr21_+_45717930 | 1.92 |
ENSDART00000164315
|
ddx46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr23_+_404975 | 1.91 |
ENSDART00000181336
|
CU929146.1
|
|
| chr2_+_19578446 | 1.89 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr8_+_48484455 | 1.88 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr13_+_2625150 | 1.88 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr8_+_8936912 | 1.88 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr8_+_30112655 | 1.86 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr22_+_10660140 | 1.84 |
ENSDART00000105835
ENSDART00000038511 |
tusc2b
|
tumor suppressor candidate 2b |
| chr10_+_40774215 | 1.83 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr21_-_45077429 | 1.82 |
ENSDART00000187268
ENSDART00000191003 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr22_+_14836040 | 1.82 |
ENSDART00000180951
|
gtpbp1l
|
GTP binding protein 1, like |
| chr12_+_23912074 | 1.80 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr25_+_2415131 | 1.78 |
ENSDART00000162210
|
zmp:0000000932
|
zmp:0000000932 |
| chr19_+_1688727 | 1.76 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr17_-_45386546 | 1.75 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr6_-_35046735 | 1.75 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr15_+_36309070 | 1.71 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr3_+_46635527 | 1.71 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr15_+_9053059 | 1.71 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr13_+_27232848 | 1.68 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr5_-_8907819 | 1.67 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr3_-_16719244 | 1.60 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chrM_+_9735 | 1.55 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr24_+_12835935 | 1.55 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr21_+_30746348 | 1.54 |
ENSDART00000050172
|
trpc2b
|
transient receptor potential cation channel, subfamily C, member 2b |
| chr23_-_31913069 | 1.53 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr14_+_45675306 | 1.50 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr7_+_38509333 | 1.47 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr21_+_25777425 | 1.42 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr7_+_756942 | 1.40 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr11_+_39107131 | 1.38 |
ENSDART00000105140
|
zgc:112255
|
zgc:112255 |
| chr10_-_25217347 | 1.38 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr4_-_9891874 | 1.33 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr10_+_6884627 | 1.33 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr4_-_16833518 | 1.31 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr7_+_69653981 | 1.26 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr14_+_34490445 | 1.20 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr21_+_43404945 | 1.19 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr3_+_13624815 | 1.18 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr21_+_4313039 | 1.13 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr9_+_54695981 | 1.10 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr2_-_59285085 | 1.10 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr16_-_43344859 | 1.08 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr16_+_13818743 | 1.07 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr20_+_39457598 | 1.04 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr7_-_17816175 | 1.01 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr20_+_12830448 | 1.01 |
ENSDART00000164754
|
BX547930.4
|
|
| chr16_+_31804590 | 0.98 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr24_+_1023839 | 0.96 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr17_-_23416897 | 0.95 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr8_+_45334255 | 0.92 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr4_+_9177997 | 0.90 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr20_-_47188966 | 0.89 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr22_+_35275468 | 0.89 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr18_-_43884044 | 0.89 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr3_-_58046319 | 0.84 |
ENSDART00000155968
|
si:ch211-256e16.10
|
si:ch211-256e16.10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 26.3 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 3.5 | 10.4 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 2.4 | 7.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 2.0 | 8.0 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 1.7 | 9.9 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 1.1 | 7.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 4.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.8 | 8.3 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.8 | 2.4 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.7 | 5.4 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.6 | 3.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.6 | 4.5 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.6 | 1.9 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.6 | 4.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.6 | 6.5 | GO:0061333 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.6 | 13.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.5 | 2.5 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.5 | 11.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.5 | 3.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.4 | 3.5 | GO:0009217 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.4 | 1.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.4 | 20.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 1.2 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.4 | 1.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.4 | 1.2 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.4 | 1.8 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.4 | 5.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 7.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.3 | 2.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.3 | 4.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.3 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 0.6 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 2.5 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.3 | 3.3 | GO:0070301 | necrotic cell death(GO:0070265) cellular response to hydrogen peroxide(GO:0070301) programmed necrotic cell death(GO:0097300) |
| 0.3 | 0.9 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.3 | 1.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 4.5 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.3 | 7.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.3 | 2.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 2.8 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 8.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 1.7 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 5.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 5.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 2.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 2.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 3.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.2 | 8.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 6.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 0.8 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.2 | 1.1 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 3.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.7 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 2.1 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 2.9 | GO:0060173 | limb development(GO:0060173) |
| 0.1 | 1.9 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 2.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.5 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 3.4 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 4.4 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 3.8 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 9.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 4.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 3.2 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.1 | 1.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 4.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 2.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 2.0 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 1.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.0 | GO:0051784 | negative regulation of mitotic nuclear division(GO:0045839) negative regulation of nuclear division(GO:0051784) |
| 0.0 | 7.4 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.6 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.6 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 2.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 2.1 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.4 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 7.5 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 2.5 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 1.3 | GO:0033334 | fin morphogenesis(GO:0033334) appendage morphogenesis(GO:0035107) |
| 0.0 | 9.2 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 1.9 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 5.6 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 4.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.1 | 7.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.8 | 4.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.7 | 2.0 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.5 | 20.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 5.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 1.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 6.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 16.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 2.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 3.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 0.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 4.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 8.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 12.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 1.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 12.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 3.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.3 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 1.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 7.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 2.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 6.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 6.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 5.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 8.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 10.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 7.3 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 13.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 13.9 | GO:0005829 | cytosol(GO:0005829) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 26.3 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 1.8 | 8.9 | GO:0009374 | biotin binding(GO:0009374) |
| 1.3 | 6.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 1.3 | 3.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.8 | 2.5 | GO:0000035 | acyl binding(GO:0000035) |
| 0.8 | 8.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.8 | 2.4 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.7 | 8.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.6 | 3.5 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.5 | 4.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.4 | 5.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 5.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 1.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 1.7 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.3 | 4.0 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 5.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 5.1 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 0.6 | GO:0032356 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 3.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 5.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 8.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 4.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.6 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 3.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 4.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 6.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 14.9 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 7.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.8 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.8 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 15.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.9 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 5.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 2.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 11.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 4.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 9.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 6.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 7.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 3.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 7.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 6.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 3.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 9.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.2 | GO:0015293 | symporter activity(GO:0015293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.4 | 8.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 7.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 3.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 14.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 8.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 8.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 3.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 2.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 16.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.7 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |