Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
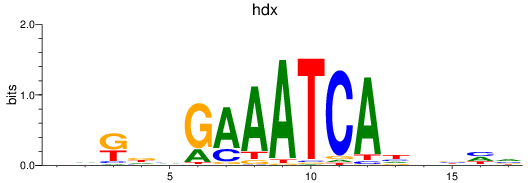
Results for hdx
Z-value: 0.65
Transcription factors associated with hdx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hdx
|
ENSDARG00000079382 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hdx | dr11_v1_chr14_-_6225336_6225336 | -0.41 | 3.8e-05 | Click! |
Activity profile of hdx motif
Sorted Z-values of hdx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_6742287 | 5.82 |
ENSDART00000140752
|
miox
|
myo-inositol oxygenase |
| chr1_+_24382651 | 5.52 |
ENSDART00000123789
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr13_-_9895564 | 5.29 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr23_-_12788113 | 4.89 |
ENSDART00000103231
ENSDART00000146640 ENSDART00000180549 |
si:dkey-96f10.1
|
si:dkey-96f10.1 |
| chr4_+_1530287 | 4.50 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr8_-_33568964 | 4.05 |
ENSDART00000138921
|
mvb12bb
|
multivesicular body subunit 12Bb |
| chr7_-_24022340 | 3.95 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr23_+_5108374 | 3.75 |
ENSDART00000114263
|
zgc:194242
|
zgc:194242 |
| chr22_-_278328 | 3.25 |
ENSDART00000098072
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr3_-_3413669 | 3.24 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr14_-_25599002 | 2.93 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr25_+_10811551 | 2.87 |
ENSDART00000167730
|
anpepb
|
alanyl (membrane) aminopeptidase b |
| chr14_-_30390145 | 2.78 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr17_-_2596125 | 2.74 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr13_+_33688474 | 2.61 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr20_-_25542183 | 2.42 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr4_-_8040436 | 2.32 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr9_-_12888082 | 2.26 |
ENSDART00000133135
ENSDART00000134415 |
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr2_-_51634431 | 2.22 |
ENSDART00000165568
|
pigr
|
polymeric immunoglobulin receptor |
| chr7_+_20031202 | 2.09 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr15_-_6946286 | 2.05 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr5_+_10046643 | 1.93 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr18_-_25276932 | 1.93 |
ENSDART00000076183
|
plin1
|
perilipin 1 |
| chr17_-_2834764 | 1.90 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr2_+_31600661 | 1.80 |
ENSDART00000139039
|
si:ch211-106h4.4
|
si:ch211-106h4.4 |
| chr8_+_8468139 | 1.70 |
ENSDART00000186684
|
comta
|
catechol-O-methyltransferase a |
| chr18_+_9810642 | 1.64 |
ENSDART00000143965
|
CACNA2D1
|
si:dkey-266j7.2 |
| chr23_+_36106790 | 1.62 |
ENSDART00000128533
|
hoxc3a
|
homeobox C3a |
| chr5_-_3885727 | 1.55 |
ENSDART00000143250
|
mlxipl
|
MLX interacting protein like |
| chr23_+_19655301 | 1.54 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr17_+_15433518 | 1.48 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr7_+_20030888 | 1.47 |
ENSDART00000192808
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr17_-_6536305 | 1.44 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr17_-_6618574 | 1.43 |
ENSDART00000184486
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr14_-_30490465 | 1.41 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr6_-_2133737 | 1.41 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr11_+_44503774 | 1.41 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr16_-_33930060 | 1.37 |
ENSDART00000110743
ENSDART00000101898 |
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr22_-_23267893 | 1.37 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr23_-_31633201 | 1.37 |
ENSDART00000143335
ENSDART00000053531 |
slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr22_+_28995391 | 1.33 |
ENSDART00000164486
|
pimr99
|
Pim proto-oncogene, serine/threonine kinase, related 99 |
| chr19_-_20270178 | 1.32 |
ENSDART00000144891
ENSDART00000090883 |
gpnmb
|
glycoprotein (transmembrane) nmb |
| chr19_+_41173386 | 1.30 |
ENSDART00000142773
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr7_+_58699718 | 1.29 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr17_-_6641535 | 1.28 |
ENSDART00000154540
ENSDART00000180384 |
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr23_+_36122058 | 1.28 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr25_-_23583101 | 1.28 |
ENSDART00000149107
ENSDART00000103704 ENSDART00000184903 |
nap1l4a
|
nucleosome assembly protein 1-like 4a |
| chr14_-_30490763 | 1.26 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr11_+_6116503 | 1.26 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr21_-_39931285 | 1.25 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr19_-_24555935 | 1.25 |
ENSDART00000132660
ENSDART00000162801 |
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr19_-_2420990 | 1.24 |
ENSDART00000181498
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr17_-_29192987 | 1.24 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr14_+_26719691 | 1.23 |
ENSDART00000078522
ENSDART00000172927 |
eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr3_+_3099492 | 1.23 |
ENSDART00000154156
|
si:dkey-30g5.1
|
si:dkey-30g5.1 |
| chr4_+_34599955 | 1.19 |
ENSDART00000150790
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr9_-_23152092 | 1.19 |
ENSDART00000180155
ENSDART00000186935 |
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr13_-_35459928 | 1.17 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr22_+_1615691 | 1.16 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr21_-_2322102 | 1.14 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr3_+_16724614 | 1.14 |
ENSDART00000182135
|
gys1
|
glycogen synthase 1 (muscle) |
| chr15_+_31428332 | 1.10 |
ENSDART00000141429
|
or103-5
|
odorant receptor, family C, subfamily 103, member 5 |
| chr17_+_48164536 | 1.10 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr21_+_21679086 | 1.09 |
ENSDART00000146225
|
or125-5
|
odorant receptor, family E, subfamily 125, member 5 |
| chr10_-_4980150 | 1.09 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr17_-_40110782 | 1.08 |
ENSDART00000126929
|
si:dkey-187k19.2
|
si:dkey-187k19.2 |
| chr3_-_40945710 | 1.08 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr8_-_48675152 | 1.07 |
ENSDART00000164544
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr2_-_24488652 | 1.07 |
ENSDART00000052067
|
insl3
|
insulin-like 3 (Leydig cell) |
| chr17_-_6536466 | 1.07 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr18_-_17077596 | 1.06 |
ENSDART00000111821
|
il17c
|
interleukin 17c |
| chr23_-_44226556 | 1.06 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr8_+_7033049 | 1.05 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr8_+_999421 | 1.04 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr15_-_18232712 | 1.02 |
ENSDART00000081199
|
wu:fj20b03
|
wu:fj20b03 |
| chr16_-_19026414 | 0.98 |
ENSDART00000141208
|
pimr98
|
Pim proto-oncogene, serine/threonine kinase, related 98 |
| chr8_-_48675411 | 0.98 |
ENSDART00000165081
|
pimr183
|
Pim proto-oncogene, serine/threonine kinase, related 183 |
| chr20_+_14789148 | 0.98 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr22_+_29027037 | 0.97 |
ENSDART00000170071
|
pimr97
|
Pim proto-oncogene, serine/threonine kinase, related 97 |
| chr6_+_21005725 | 0.97 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr3_+_25154078 | 0.96 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr8_-_53044089 | 0.96 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr5_-_4245186 | 0.96 |
ENSDART00000145912
|
si:ch211-283g2.3
|
si:ch211-283g2.3 |
| chr14_+_34495216 | 0.95 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr25_-_18778356 | 0.95 |
ENSDART00000111425
|
fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr11_-_29737088 | 0.94 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr2_-_40890004 | 0.93 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr23_+_37579107 | 0.92 |
ENSDART00000169376
|
plekhg5b
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5b |
| chr20_+_6543674 | 0.92 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr17_+_6536152 | 0.92 |
ENSDART00000062952
ENSDART00000121789 |
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr22_-_5171362 | 0.90 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr10_-_36721726 | 0.90 |
ENSDART00000122132
|
pimr137
|
Pim proto-oncogene, serine/threonine kinase, related 137 |
| chr4_-_64141714 | 0.89 |
ENSDART00000128628
|
BX914205.3
|
|
| chr8_+_17884569 | 0.88 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr25_+_19710619 | 0.87 |
ENSDART00000156435
|
mxg
|
myxovirus (influenza virus) resistance G |
| chr10_-_25570647 | 0.86 |
ENSDART00000134215
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr6_-_9459440 | 0.86 |
ENSDART00000064991
|
pimr71
|
Pim proto-oncogene, serine/threonine kinase, related 71 |
| chr15_+_25683069 | 0.85 |
ENSDART00000148190
|
hic1
|
hypermethylated in cancer 1 |
| chr7_+_22399669 | 0.84 |
ENSDART00000144375
|
fgf11a
|
fibroblast growth factor 11a |
| chr8_-_53044300 | 0.84 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr19_-_18626952 | 0.83 |
ENSDART00000168004
ENSDART00000162034 ENSDART00000165486 ENSDART00000167971 |
rps18
|
ribosomal protein S18 |
| chr6_-_24143923 | 0.83 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr5_+_27267186 | 0.82 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr22_-_4944795 | 0.81 |
ENSDART00000147116
|
pku300
|
pku300 |
| chr20_+_10727022 | 0.81 |
ENSDART00000104185
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr16_+_26735894 | 0.80 |
ENSDART00000191605
ENSDART00000185175 |
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr4_+_70556298 | 0.80 |
ENSDART00000170985
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr6_+_72040 | 0.80 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr18_+_41820039 | 0.79 |
ENSDART00000128345
|
pgr
|
progesterone receptor |
| chr25_-_15268443 | 0.79 |
ENSDART00000151827
|
ccdc73
|
coiled-coil domain containing 73 |
| chr6_+_16982613 | 0.79 |
ENSDART00000156104
|
pimr8
|
Pim proto-oncogene, serine/threonine kinase, related 8 |
| chr15_-_30989360 | 0.79 |
ENSDART00000155265
|
lgals9l6
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 6 |
| chr5_-_30620625 | 0.78 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr16_+_38820486 | 0.78 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr15_-_22304193 | 0.78 |
ENSDART00000115158
|
zgc:194114
|
zgc:194114 |
| chr5_+_62356304 | 0.76 |
ENSDART00000148381
|
aspa
|
aspartoacylase |
| chr4_-_43070566 | 0.76 |
ENSDART00000183779
|
si:dkey-54j5.2
|
si:dkey-54j5.2 |
| chr25_+_29160102 | 0.74 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr4_-_51692811 | 0.74 |
ENSDART00000159625
|
si:dkeyp-90h9.1
|
si:dkeyp-90h9.1 |
| chr3_+_43955864 | 0.74 |
ENSDART00000168267
|
BX571715.1
|
|
| chr9_-_5318873 | 0.73 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr12_+_13118540 | 0.73 |
ENSDART00000077840
ENSDART00000127870 |
cmn
|
calymmin |
| chr8_-_13428740 | 0.72 |
ENSDART00000131826
|
CR354547.3
|
|
| chr1_-_11519934 | 0.72 |
ENSDART00000162060
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr23_+_17437554 | 0.72 |
ENSDART00000184282
|
BX649300.3
|
|
| chr4_-_10958074 | 0.72 |
ENSDART00000150478
|
si:ch211-161n3.4
|
si:ch211-161n3.4 |
| chr25_+_36333993 | 0.71 |
ENSDART00000184379
|
CR354435.2
|
|
| chr16_-_26820634 | 0.71 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr16_-_54471235 | 0.71 |
ENSDART00000061572
|
cyp11c1
|
cytochrome P450, family 11, subfamily C, polypeptide 1 |
| chr4_-_10599062 | 0.71 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr21_-_28439596 | 0.71 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr18_+_15706160 | 0.71 |
ENSDART00000131524
|
si:ch211-264e16.1
|
si:ch211-264e16.1 |
| chr10_-_18492617 | 0.70 |
ENSDART00000179936
ENSDART00000193799 ENSDART00000193198 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr11_-_25775447 | 0.70 |
ENSDART00000161301
|
plch2b
|
phospholipase C, eta 2b |
| chr3_-_33029938 | 0.69 |
ENSDART00000181730
|
CT583642.1
|
|
| chr24_+_9881219 | 0.69 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr9_-_43073960 | 0.69 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr4_+_2269815 | 0.69 |
ENSDART00000075788
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr23_+_43870886 | 0.68 |
ENSDART00000102658
ENSDART00000149088 |
nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr1_+_8579197 | 0.68 |
ENSDART00000186329
|
AL928650.4
|
|
| chr2_-_36494308 | 0.67 |
ENSDART00000110378
|
BX901889.2
|
|
| chr2_+_45696743 | 0.67 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr10_+_21722892 | 0.66 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr9_+_25840720 | 0.66 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr2_-_40890264 | 0.66 |
ENSDART00000123886
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr7_-_29223614 | 0.66 |
ENSDART00000173598
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr19_-_43657468 | 0.65 |
ENSDART00000150940
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr2_-_36497113 | 0.65 |
ENSDART00000109291
|
BX901889.4
|
|
| chr4_-_16706776 | 0.64 |
ENSDART00000079461
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr1_-_28950366 | 0.64 |
ENSDART00000110270
|
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr13_-_48295723 | 0.64 |
ENSDART00000115137
|
KCNK12
|
potassium two pore domain channel subfamily K member 12 |
| chr8_+_12975963 | 0.64 |
ENSDART00000132821
ENSDART00000181755 ENSDART00000185061 ENSDART00000193727 |
dram2a
|
DNA-damage regulated autophagy modulator 2a |
| chr17_-_6535941 | 0.64 |
ENSDART00000109249
|
cenpo
|
centromere protein O |
| chr22_-_4784658 | 0.64 |
ENSDART00000140839
|
CU570881.1
|
|
| chr11_+_6752468 | 0.64 |
ENSDART00000187046
ENSDART00000182089 |
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr24_-_27409599 | 0.63 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr6_-_16868834 | 0.62 |
ENSDART00000156503
|
pimr41
|
Pim proto-oncogene, serine/threonine kinase, related 41 |
| chr8_-_48770163 | 0.61 |
ENSDART00000160959
|
pimr184
|
Pim proto-oncogene, serine/threonine kinase, related 184 |
| chr2_-_40196547 | 0.59 |
ENSDART00000168098
|
ccl34a.3
|
chemokine (C-C motif) ligand 34a, duplicate 3 |
| chr13_+_40686133 | 0.59 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr21_+_17051478 | 0.59 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr25_+_22587306 | 0.58 |
ENSDART00000067479
|
stra6
|
stimulated by retinoic acid 6 |
| chr16_+_4181105 | 0.57 |
ENSDART00000180921
|
yrdc
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr5_-_37247654 | 0.56 |
ENSDART00000112312
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr4_+_391297 | 0.56 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr11_+_5565082 | 0.55 |
ENSDART00000112590
ENSDART00000183207 |
si:ch73-40i7.5
|
si:ch73-40i7.5 |
| chr20_-_25644131 | 0.55 |
ENSDART00000138997
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr18_-_17077419 | 0.54 |
ENSDART00000148714
|
il17c
|
interleukin 17c |
| chr14_+_15620640 | 0.54 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr14_+_46287296 | 0.53 |
ENSDART00000183620
|
cabp2b
|
calcium binding protein 2b |
| chr7_-_19614916 | 0.53 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr4_-_16706941 | 0.52 |
ENSDART00000033053
ENSDART00000140510 ENSDART00000141119 |
dennd5b
|
DENN/MADD domain containing 5B |
| chr9_-_12886108 | 0.52 |
ENSDART00000177283
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr17_+_6535694 | 0.51 |
ENSDART00000191681
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr4_-_29753895 | 0.51 |
ENSDART00000150742
|
si:dkey-191j3.1
|
si:dkey-191j3.1 |
| chr9_+_8364553 | 0.50 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr4_+_7508316 | 0.50 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr11_+_30295582 | 0.50 |
ENSDART00000122424
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr7_+_49865049 | 0.50 |
ENSDART00000164165
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr12_+_30788912 | 0.50 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr21_-_32684570 | 0.49 |
ENSDART00000162873
|
adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr23_+_37815645 | 0.49 |
ENSDART00000011627
|
irx7
|
iroquois homeobox 7 |
| chr25_+_3564220 | 0.49 |
ENSDART00000170005
|
zgc:194202
|
zgc:194202 |
| chr7_-_1148539 | 0.48 |
ENSDART00000173447
|
si:dkey-31g16.2
|
si:dkey-31g16.2 |
| chr9_+_22080122 | 0.48 |
ENSDART00000065956
ENSDART00000136014 |
crygm2e
|
crystallin, gamma M2e |
| chr7_+_19615056 | 0.47 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr18_-_7112882 | 0.47 |
ENSDART00000176292
|
si:dkey-88e18.8
|
si:dkey-88e18.8 |
| chr2_+_25378457 | 0.46 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr6_+_49082796 | 0.46 |
ENSDART00000182446
|
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr13_+_38213694 | 0.46 |
ENSDART00000057320
|
zgc:171579
|
zgc:171579 |
| chr3_+_5504990 | 0.45 |
ENSDART00000143626
|
si:ch73-264i18.2
|
si:ch73-264i18.2 |
| chr7_+_17353030 | 0.45 |
ENSDART00000098020
|
si:busm1-169i8.11
|
si:busm1-169i8.11 |
| chr4_-_71108793 | 0.45 |
ENSDART00000189856
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr1_-_29761086 | 0.44 |
ENSDART00000136760
|
alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr7_-_17268111 | 0.44 |
ENSDART00000169319
|
nitr10a
|
novel immune-type receptor 10a |
| chr25_-_21609645 | 0.43 |
ENSDART00000087817
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr20_-_26841544 | 0.43 |
ENSDART00000088875
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr18_+_814328 | 0.42 |
ENSDART00000157801
ENSDART00000188492 ENSDART00000184489 |
fam219b
|
family with sequence similarity 219, member B |
| chr14_+_12316581 | 0.42 |
ENSDART00000170115
ENSDART00000149757 |
si:ch211-125c23.3
cx31.7
|
si:ch211-125c23.3 connexin 31.7 |
| chr10_+_39164638 | 0.40 |
ENSDART00000188997
|
CU633908.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hdx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 1.0 | 4.0 | GO:1902592 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.7 | 2.9 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.6 | 5.5 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.5 | 2.8 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.4 | 5.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.4 | 1.4 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 0.9 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 0.8 | GO:2000793 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.3 | 1.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 4.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.7 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.2 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 0.7 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.2 | 1.4 | GO:0046323 | glucose import(GO:0046323) |
| 0.2 | 2.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.8 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 2.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.6 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.1 | 0.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 2.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 4.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 3.7 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 1.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.7 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 1.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.3 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 1.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 2.4 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.7 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 3.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 1.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 1.8 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 1.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 8.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.9 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 2.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.2 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 1.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.7 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 3.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.8 | 3.1 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.3 | 0.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 3.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 2.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 0.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 0.6 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.7 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.3 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 3.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.7 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.9 | 3.7 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.7 | 2.9 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.5 | 5.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 1.7 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 5.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 2.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 1.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 1.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 2.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 4.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 1.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 1.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 1.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 2.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 2.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.8 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.7 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 1.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.7 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 3.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 4.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 3.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 3.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 2.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 1.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |