Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
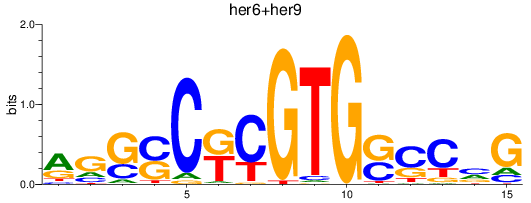
Results for her6+her9
Z-value: 0.46
Transcription factors associated with her6+her9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
her6
|
ENSDARG00000006514 | hairy-related 6 |
|
her9
|
ENSDARG00000056438 | hairy-related 9 |
|
her6
|
ENSDARG00000111099 | hairy-related 6 |
|
her9
|
ENSDARG00000115775 | hairy-related 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| her6 | dr11_v1_chr6_-_36552844_36552844 | 0.53 | 4.1e-08 | Click! |
| her9 | dr11_v1_chr23_-_23401305_23401305 | 0.53 | 4.4e-08 | Click! |
Activity profile of her6+her9 motif
Sorted Z-values of her6+her9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77551860 | 3.63 |
ENSDART00000188176
|
AL935186.6
|
|
| chr4_-_77563411 | 3.41 |
ENSDART00000186841
|
AL935186.8
|
|
| chr24_-_33291784 | 3.26 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr2_+_68789 | 3.24 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr24_+_23791758 | 2.65 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr2_-_58257624 | 2.58 |
ENSDART00000098940
|
foxl2b
|
forkhead box L2b |
| chr23_+_22656477 | 2.51 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr8_-_912821 | 2.40 |
ENSDART00000082296
|
ptger4a
|
prostaglandin E receptor 4 (subtype EP4) a |
| chr4_+_68562464 | 2.17 |
ENSDART00000192954
|
BX548011.4
|
|
| chr22_-_10502780 | 2.13 |
ENSDART00000136961
|
ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr12_+_48340133 | 2.01 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr10_-_2942900 | 1.90 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr20_+_39283849 | 1.80 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr19_+_19777437 | 1.74 |
ENSDART00000170662
|
hoxa3a
|
homeobox A3a |
| chr22_-_193234 | 1.65 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr18_+_5490668 | 1.64 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr22_+_19311411 | 1.34 |
ENSDART00000133234
ENSDART00000138284 |
si:dkey-21e2.16
|
si:dkey-21e2.16 |
| chr5_+_35398745 | 1.05 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr1_-_38908197 | 1.01 |
ENSDART00000101489
|
vegfc
|
vascular endothelial growth factor c |
| chr1_+_31942961 | 1.00 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr3_-_8765165 | 0.96 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr23_-_2081554 | 0.92 |
ENSDART00000179805
|
ndnf
|
neuron-derived neurotrophic factor |
| chr4_+_9669717 | 0.91 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr19_+_19786117 | 0.89 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr18_-_494606 | 0.88 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr2_+_58841181 | 0.80 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr19_-_35450661 | 0.78 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr8_+_12925385 | 0.75 |
ENSDART00000085377
|
KIF2A
|
zgc:103670 |
| chr6_-_40195510 | 0.61 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr13_-_45022527 | 0.59 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr5_-_1963498 | 0.59 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr18_+_16125852 | 0.53 |
ENSDART00000061106
|
bhlhe41
|
basic helix-loop-helix family, member e41 |
| chr23_-_9919959 | 0.53 |
ENSDART00000127029
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr4_+_62576422 | 0.52 |
ENSDART00000180679
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr23_+_36616717 | 0.51 |
ENSDART00000042701
ENSDART00000192980 |
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr3_-_58823762 | 0.51 |
ENSDART00000101253
|
luc7l3
|
LUC7-like 3 pre-mRNA splicing factor |
| chr9_-_43538328 | 0.49 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr8_-_39977026 | 0.49 |
ENSDART00000141707
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr1_-_54107321 | 0.45 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr23_+_44732863 | 0.44 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr13_-_45022301 | 0.42 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr9_+_9112159 | 0.38 |
ENSDART00000165932
|
sik1
|
salt-inducible kinase 1 |
| chr24_+_26432541 | 0.35 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr5_-_32890807 | 0.35 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr5_-_36916790 | 0.31 |
ENSDART00000143827
|
kptn
|
kaptin (actin binding protein) |
| chr10_-_1788376 | 0.30 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_+_13069168 | 0.29 |
ENSDART00000192832
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr15_-_12319065 | 0.26 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr13_+_7241170 | 0.25 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr5_-_23429228 | 0.25 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr16_+_11029762 | 0.23 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr20_-_40766387 | 0.22 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr9_+_30213144 | 0.20 |
ENSDART00000139920
|
senp7a
|
SUMO1/sentrin specific peptidase 7a |
| chr3_-_11828206 | 0.19 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr5_-_12560569 | 0.19 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr19_-_6385594 | 0.16 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr7_+_24528430 | 0.13 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr3_+_14641962 | 0.09 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr11_-_12800945 | 0.01 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr11_-_12801157 | 0.01 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr23_+_40452157 | 0.01 |
ENSDART00000113106
ENSDART00000140136 |
soga3b
|
SOGA family member 3b |
| chr20_-_5328637 | 0.01 |
ENSDART00000075974
|
ism2b
|
isthmin 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of her6+her9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.7 | 3.4 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.3 | 2.5 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 0.9 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 1.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.9 | GO:0021543 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.2 | 0.8 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 3.3 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 1.0 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 5.5 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 2.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 1.0 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 2.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 5.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.5 | 2.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 3.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 1.0 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.3 | 2.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 1.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.2 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |