Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
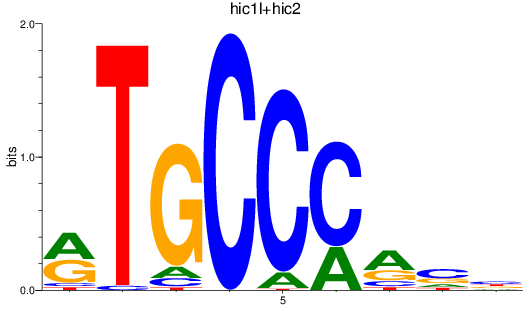
Results for hic1l+hic2
Z-value: 1.03
Transcription factors associated with hic1l+hic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hic1l
|
ENSDARG00000045660 | hypermethylated in cancer 1 like |
|
hic2
|
ENSDARG00000100497 | hypermethylated in cancer 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hic1l | dr11_v1_chr25_-_25575717_25575717 | -0.80 | 9.4e-22 | Click! |
| hic2 | dr11_v1_chr10_+_3145707_3145707 | -0.61 | 8.7e-11 | Click! |
Activity profile of hic1l+hic2 motif
Sorted Z-values of hic1l+hic2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_25612680 | 21.57 |
ENSDART00000114167
|
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr7_+_29952719 | 18.70 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr7_+_29952169 | 18.53 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr14_-_21064199 | 12.84 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr1_-_55810730 | 9.76 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr2_-_44255537 | 9.18 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr18_-_16123222 | 7.93 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr3_+_24207243 | 7.87 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr16_+_12836143 | 7.87 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr25_-_31423493 | 7.67 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr3_+_23221047 | 7.28 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr19_+_14109348 | 7.18 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr5_-_57641257 | 6.70 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr18_-_50845804 | 5.66 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr10_+_15608326 | 5.31 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr5_+_32815745 | 5.20 |
ENSDART00000181535
|
crata
|
carnitine O-acetyltransferase a |
| chr4_-_77551860 | 5.03 |
ENSDART00000188176
|
AL935186.6
|
|
| chr24_-_29822913 | 5.02 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr14_-_21063977 | 4.82 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr8_+_38417461 | 4.79 |
ENSDART00000132718
|
nkx6.3
|
NK6 homeobox 3 |
| chr13_-_7573670 | 4.67 |
ENSDART00000102538
|
pitx3
|
paired-like homeodomain 3 |
| chr6_+_29402997 | 4.64 |
ENSDART00000104298
|
ndufb5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 |
| chr3_+_23692462 | 4.43 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr4_-_8030583 | 4.23 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr24_-_9989634 | 4.16 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr2_+_24936766 | 4.11 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr14_+_6159162 | 4.05 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr5_-_69041102 | 3.77 |
ENSDART00000161561
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr4_-_14915268 | 3.74 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr15_+_25158104 | 3.14 |
ENSDART00000128267
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr21_+_5257018 | 3.08 |
ENSDART00000183100
ENSDART00000191525 |
loxhd1a
|
lipoxygenase homology domains 1a |
| chr11_-_16021424 | 3.07 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr23_-_27875140 | 2.97 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr8_+_15254564 | 2.93 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr21_-_22681534 | 2.89 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr4_-_8040436 | 2.89 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr10_-_8129175 | 2.88 |
ENSDART00000133921
|
ndufa9b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9b |
| chr10_+_43188678 | 2.70 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr4_-_25485404 | 2.67 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr25_+_16915974 | 2.59 |
ENSDART00000188923
|
zgc:77158
|
zgc:77158 |
| chr23_-_1587955 | 2.57 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr20_+_27003713 | 2.56 |
ENSDART00000153232
|
si:dkey-177p2.18
|
si:dkey-177p2.18 |
| chr3_-_32170850 | 2.44 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr6_-_29402578 | 2.44 |
ENSDART00000115031
|
mrpl47
|
mitochondrial ribosomal protein L47 |
| chr2_-_37889111 | 2.37 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr21_-_42876565 | 2.32 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr15_-_37779978 | 2.30 |
ENSDART00000157202
|
si:dkey-42l23.3
|
si:dkey-42l23.3 |
| chr1_+_30723380 | 2.29 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr14_-_897874 | 2.26 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr25_+_36292465 | 2.25 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr2_+_30547018 | 2.21 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr15_-_17138640 | 2.07 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr23_-_31060350 | 2.06 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr5_-_28679135 | 2.04 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr7_+_16963091 | 2.00 |
ENSDART00000173770
|
nav2a
|
neuron navigator 2a |
| chr4_-_5597167 | 2.00 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr24_+_34806822 | 1.96 |
ENSDART00000148407
ENSDART00000188328 |
mchr2
|
melanin-concentrating hormone receptor 2 |
| chr6_-_49634787 | 1.95 |
ENSDART00000188538
|
BX323459.1
|
|
| chr8_-_14144707 | 1.94 |
ENSDART00000148061
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr11_+_12879635 | 1.94 |
ENSDART00000182515
ENSDART00000081296 |
si:dkey-11m19.5
|
si:dkey-11m19.5 |
| chr13_+_31205439 | 1.93 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr13_-_9061944 | 1.87 |
ENSDART00000164186
ENSDART00000102051 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr6_+_13726844 | 1.82 |
ENSDART00000055833
|
wnt6a
|
wingless-type MMTV integration site family, member 6a |
| chr25_+_19681662 | 1.81 |
ENSDART00000104347
|
si:dkeyp-110c12.3
|
si:dkeyp-110c12.3 |
| chr23_-_32404022 | 1.81 |
ENSDART00000156387
ENSDART00000155508 |
si:ch211-66i15.4
|
si:ch211-66i15.4 |
| chr19_+_17642356 | 1.80 |
ENSDART00000176431
|
CR382334.1
|
|
| chr2_-_25159309 | 1.77 |
ENSDART00000137290
|
si:dkey-223d7.6
|
si:dkey-223d7.6 |
| chr25_-_3087556 | 1.73 |
ENSDART00000193249
|
best1
|
bestrophin 1 |
| chr11_+_13032483 | 1.72 |
ENSDART00000108968
|
ptgfr
|
prostaglandin F receptor (FP) |
| chr8_-_38616712 | 1.72 |
ENSDART00000141827
|
si:ch211-198d23.1
|
si:ch211-198d23.1 |
| chr22_+_7497319 | 1.71 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr2_+_17181777 | 1.71 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr16_-_25606889 | 1.69 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr16_-_41840668 | 1.67 |
ENSDART00000146150
|
si:dkey-199f5.4
|
si:dkey-199f5.4 |
| chr21_+_13244450 | 1.66 |
ENSDART00000146062
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr7_-_65236649 | 1.65 |
ENSDART00000185070
|
pkd1l2a
|
polycystic kidney disease 1 like 2a |
| chr8_-_53548145 | 1.62 |
ENSDART00000180665
|
parapinopsina
|
parapinopsin a |
| chr20_+_23185530 | 1.62 |
ENSDART00000142346
|
lrrc66
|
leucine rich repeat containing 66 |
| chr6_+_40723554 | 1.61 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr25_-_7686201 | 1.61 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr8_+_46536893 | 1.60 |
ENSDART00000124023
|
pimr187
|
Pim proto-oncogene, serine/threonine kinase, related 187 |
| chr9_-_19610436 | 1.59 |
ENSDART00000108697
|
fgf9
|
fibroblast growth factor 9 |
| chr23_-_1144425 | 1.58 |
ENSDART00000098271
|
psmb11b
|
proteasome subunit beta11b |
| chr21_+_25375172 | 1.58 |
ENSDART00000078748
|
or132-5
|
odorant receptor, family H, subfamily 132, member 5 |
| chr23_-_2037566 | 1.57 |
ENSDART00000191312
ENSDART00000127443 |
prdm5
|
PR domain containing 5 |
| chr1_+_44992207 | 1.56 |
ENSDART00000172357
|
tmem132a
|
transmembrane protein 132A |
| chr6_-_57655030 | 1.53 |
ENSDART00000155438
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr22_+_29009541 | 1.53 |
ENSDART00000169449
|
pimr97
|
Pim proto-oncogene, serine/threonine kinase, related 97 |
| chr8_-_7475917 | 1.52 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr3_-_1387292 | 1.51 |
ENSDART00000163535
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr23_-_21797517 | 1.50 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr2_+_30531726 | 1.49 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr18_+_33100606 | 1.48 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr5_+_39537195 | 1.47 |
ENSDART00000051256
|
fgf5
|
fibroblast growth factor 5 |
| chr5_+_61738276 | 1.46 |
ENSDART00000186256
|
RASL10B
|
RAS like family 10 member B |
| chr25_-_16146851 | 1.44 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr2_-_30912922 | 1.41 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr14_-_448182 | 1.40 |
ENSDART00000180018
|
FAT4
|
FAT atypical cadherin 4 |
| chr13_+_32148338 | 1.40 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr1_+_30723677 | 1.38 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr16_-_41807397 | 1.38 |
ENSDART00000138577
|
si:dkey-199f5.7
|
si:dkey-199f5.7 |
| chr13_-_37383625 | 1.38 |
ENSDART00000192888
ENSDART00000100322 ENSDART00000138934 |
kcnh5b
|
potassium voltage-gated channel, subfamily H (eag-related), member 5b |
| chr15_-_37425468 | 1.37 |
ENSDART00000059630
|
si:ch211-113j13.2
|
si:ch211-113j13.2 |
| chr11_+_20057182 | 1.37 |
ENSDART00000184979
|
fezf2
|
FEZ family zinc finger 2 |
| chr22_+_30137374 | 1.33 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr8_+_18101608 | 1.32 |
ENSDART00000140941
|
glis1b
|
GLIS family zinc finger 1b |
| chr19_+_4463389 | 1.31 |
ENSDART00000168805
|
kcnk9
|
potassium channel, subfamily K, member 9 |
| chr12_-_22524388 | 1.29 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr17_+_15845765 | 1.28 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr18_+_20677090 | 1.28 |
ENSDART00000060243
|
cftr
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr21_-_26520629 | 1.26 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr22_-_5006119 | 1.23 |
ENSDART00000187998
|
rx1
|
retinal homeobox gene 1 |
| chr2_-_23600821 | 1.21 |
ENSDART00000146217
|
BX677666.1
|
|
| chr13_-_14926318 | 1.20 |
ENSDART00000142785
|
cdc25b
|
cell division cycle 25B |
| chr16_-_25606235 | 1.15 |
ENSDART00000192741
|
zgc:110410
|
zgc:110410 |
| chr1_-_54425791 | 1.11 |
ENSDART00000039911
|
pkd1a
|
polycystic kidney disease 1a |
| chr2_-_37883256 | 1.10 |
ENSDART00000035685
|
hbl4
|
hexose-binding lectin 4 |
| chr14_-_41535822 | 1.07 |
ENSDART00000149407
|
itga6l
|
integrin, alpha 6, like |
| chr12_+_28995942 | 1.06 |
ENSDART00000076334
|
valopb
|
vertebrate ancient long opsin b |
| chr7_-_38689562 | 1.05 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr22_+_7742211 | 1.04 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr21_+_18877130 | 1.04 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr5_-_10236599 | 1.03 |
ENSDART00000099834
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr17_+_53294228 | 1.02 |
ENSDART00000158172
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr13_+_21870269 | 1.01 |
ENSDART00000144612
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr5_-_57820873 | 0.98 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr20_+_48543492 | 0.96 |
ENSDART00000175480
|
LO018254.1
|
|
| chr25_+_30238021 | 0.96 |
ENSDART00000123220
|
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr4_+_54519511 | 0.95 |
ENSDART00000161653
|
znf974
|
zinc finger protein 974 |
| chr3_-_61592417 | 0.90 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr10_+_44437080 | 0.90 |
ENSDART00000181517
ENSDART00000054153 |
denr
|
density-regulated protein |
| chr20_-_25481306 | 0.89 |
ENSDART00000182495
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr15_+_42626957 | 0.86 |
ENSDART00000154754
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr2_-_41942666 | 0.85 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr13_-_45811137 | 0.85 |
ENSDART00000190512
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr15_-_7337148 | 0.83 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr7_+_26172396 | 0.83 |
ENSDART00000173975
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr1_-_34685007 | 0.80 |
ENSDART00000157471
|
klf5a
|
Kruppel-like factor 5a |
| chr13_+_36633355 | 0.79 |
ENSDART00000135612
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr17_+_52823015 | 0.79 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr9_-_41817712 | 0.78 |
ENSDART00000182657
|
si:dkeyp-30e7.2
|
si:dkeyp-30e7.2 |
| chr2_+_9536021 | 0.78 |
ENSDART00000187357
|
CU929379.1
|
|
| chr15_-_13254480 | 0.75 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr25_-_27541621 | 0.74 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr7_+_22981909 | 0.74 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr5_-_6508250 | 0.72 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr10_-_34867401 | 0.71 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr12_+_22680115 | 0.71 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr7_-_65191937 | 0.71 |
ENSDART00000173234
|
pkd1l2a
|
polycystic kidney disease 1 like 2a |
| chr6_+_8630355 | 0.69 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr7_-_30143092 | 0.68 |
ENSDART00000173636
|
frmd5
|
FERM domain containing 5 |
| chr17_-_19344999 | 0.62 |
ENSDART00000138315
|
gsc
|
goosecoid |
| chr23_+_22293682 | 0.62 |
ENSDART00000187304
|
CR354612.1
|
|
| chr16_-_53800047 | 0.61 |
ENSDART00000158047
|
znrf2b
|
zinc and ring finger 2b |
| chr11_-_6001845 | 0.59 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr12_-_20362041 | 0.58 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr8_+_36570791 | 0.56 |
ENSDART00000145566
ENSDART00000180527 |
pold2
|
polymerase (DNA directed), delta 2, regulatory subunit |
| chr8_-_1219815 | 0.55 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr15_-_28082310 | 0.54 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr12_+_5251647 | 0.54 |
ENSDART00000124097
|
plce1
|
phospholipase C, epsilon 1 |
| chr17_+_27134806 | 0.54 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr8_+_35664152 | 0.53 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr21_+_10794914 | 0.52 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr9_+_38737924 | 0.50 |
ENSDART00000147652
|
kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr10_-_12650259 | 0.48 |
ENSDART00000191567
|
CT990561.2
|
|
| chr2_+_31475772 | 0.47 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr23_-_6765653 | 0.46 |
ENSDART00000192310
|
FP102169.1
|
|
| chr12_+_35654749 | 0.44 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr11_+_4953964 | 0.44 |
ENSDART00000092552
|
ptprga
|
protein tyrosine phosphatase, receptor type, g a |
| chr8_+_8973425 | 0.42 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr9_-_33107237 | 0.42 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr10_-_26179805 | 0.42 |
ENSDART00000174797
|
trim3b
|
tripartite motif containing 3b |
| chr23_+_2669 | 0.42 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr14_+_25464681 | 0.41 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr12_-_47580562 | 0.37 |
ENSDART00000153431
|
rgs7b
|
regulator of G protein signaling 7b |
| chr3_+_9588705 | 0.37 |
ENSDART00000172543
ENSDART00000104875 |
trap1
|
TNF receptor-associated protein 1 |
| chr10_+_41199660 | 0.36 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr19_-_2115040 | 0.36 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr3_-_61065055 | 0.35 |
ENSDART00000074199
|
cacng1b
|
calcium channel, voltage-dependent, gamma subunit 1b |
| chr16_+_35594670 | 0.34 |
ENSDART00000163275
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr10_+_42691210 | 0.33 |
ENSDART00000193813
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr5_-_38121612 | 0.31 |
ENSDART00000159543
|
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr25_+_7229046 | 0.28 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr2_-_14798295 | 0.28 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr13_+_39297802 | 0.25 |
ENSDART00000133636
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr11_-_41220794 | 0.23 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr15_-_11956981 | 0.23 |
ENSDART00000164163
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr2_-_14793343 | 0.22 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr22_-_3595439 | 0.19 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr20_+_51466046 | 0.18 |
ENSDART00000178065
|
tlr5b
|
toll-like receptor 5b |
| chr7_+_22982201 | 0.18 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr13_-_33207367 | 0.17 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr21_+_21612214 | 0.17 |
ENSDART00000008099
|
b9d2
|
B9 domain containing 2 |
| chr22_+_5123479 | 0.17 |
ENSDART00000111822
ENSDART00000081910 |
loxl5a
|
lysyl oxidase-like 5a |
| chr17_+_26657896 | 0.16 |
ENSDART00000152037
|
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr6_-_426041 | 0.16 |
ENSDART00000162789
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr1_+_32791920 | 0.15 |
ENSDART00000109369
|
zgc:174320
|
zgc:174320 |
| chr12_+_31729498 | 0.07 |
ENSDART00000188546
ENSDART00000182562 ENSDART00000186147 |
RNF157
|
si:dkey-49c17.3 |
| chr5_-_54148271 | 0.07 |
ENSDART00000163034
|
arhgef15
|
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr16_+_35344031 | 0.06 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hic1l+hic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.7 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 3.3 | 9.8 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 3.1 | 9.2 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 2.6 | 7.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 1.8 | 7.3 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 1.6 | 8.2 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 1.0 | 5.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.0 | 2.9 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.9 | 7.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.7 | 2.0 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.6 | 4.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.6 | 1.7 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.5 | 1.4 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.5 | 2.3 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.4 | 1.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.4 | 1.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.4 | 5.0 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.4 | 37.2 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.3 | 2.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.6 | GO:0016037 | light absorption(GO:0016037) |
| 0.3 | 1.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.3 | 0.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 0.9 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.3 | 1.1 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.3 | 1.6 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.3 | 3.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 2.7 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 1.4 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.2 | 4.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.2 | 2.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.0 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 1.5 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.1 | 0.4 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 | 0.4 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 2.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 2.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 1.6 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 | 1.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 2.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 2.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 1.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.7 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.9 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.6 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 1.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 1.6 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.0 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.2 | 8.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 38.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 2.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 1.9 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 5.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 10.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 4.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 4.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.0 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 4.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 5.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 3.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 7.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 1.3 | 5.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 1.3 | 5.0 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 1.1 | 7.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.9 | 9.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.9 | 2.6 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.6 | 5.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.5 | 2.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 2.9 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 1.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 2.6 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 8.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 36.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.2 | 1.0 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 9.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 2.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.2 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 3.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 23.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.0 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 2.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 7.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 4.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 2.1 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 2.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 4.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.5 | 3.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.5 | 5.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 1.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 1.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 2.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 6.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 4.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 2.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |