Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for hlx1
Z-value: 0.36
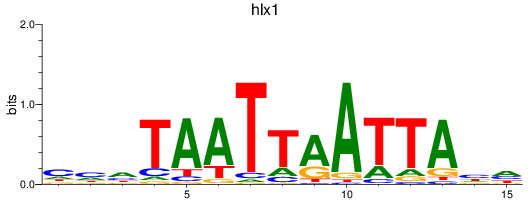
Transcription factors associated with hlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hlx1
|
ENSDARG00000009134 | H2.0-like homeo box 1 (Drosophila) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hlx1 | dr11_v1_chr20_-_52199296_52199311 | -0.57 | 2.0e-09 | Click! |
Activity profile of hlx1 motif
Sorted Z-values of hlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_22918214 | 4.82 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr3_+_29714775 | 4.81 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr2_+_47623202 | 2.91 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr5_-_66702479 | 2.85 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr17_+_9310259 | 2.59 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr1_+_6172786 | 2.58 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr3_-_48716422 | 2.57 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr3_-_20040636 | 2.56 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr10_+_26926654 | 2.49 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr1_-_26294995 | 2.33 |
ENSDART00000168594
|
cxxc4
|
CXXC finger 4 |
| chr9_-_23118350 | 2.04 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr1_-_19502322 | 1.59 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr2_+_2223837 | 1.58 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr12_-_2522487 | 1.56 |
ENSDART00000022471
ENSDART00000145213 |
mapk8b
|
mitogen-activated protein kinase 8b |
| chr8_-_39952727 | 1.15 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr24_-_21404367 | 1.10 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr10_+_43039947 | 1.09 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr13_+_22964868 | 1.05 |
ENSDART00000142129
|
tacr2
|
tachykinin receptor 2 |
| chr8_+_50953776 | 0.97 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr17_+_49281597 | 0.95 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr6_+_51773873 | 0.93 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr22_-_20950448 | 0.69 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr17_-_29119362 | 0.61 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr16_+_7380463 | 0.53 |
ENSDART00000029727
ENSDART00000149086 |
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr17_+_7620220 | 0.43 |
ENSDART00000081447
|
fbxo30a
|
F-box protein 30a |
| chr8_+_7801060 | 0.40 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr2_-_9818640 | 0.36 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr4_-_22338906 | 0.34 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr8_+_12930216 | 0.32 |
ENSDART00000115405
|
KIF2A
|
zgc:103670 |
| chr5_-_23581355 | 0.30 |
ENSDART00000191869
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr8_-_11834599 | 0.23 |
ENSDART00000190986
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr21_+_28478663 | 0.14 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr9_+_45227028 | 0.12 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 1.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.3 | 2.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.3 | 4.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 0.5 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 1.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 1.0 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 1.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.6 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 2.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 4.8 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 1.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.5 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.1 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.0 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 4.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.6 | GO:0031256 | leading edge membrane(GO:0031256) presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.3 | 2.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 2.6 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 4.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 4.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |