Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
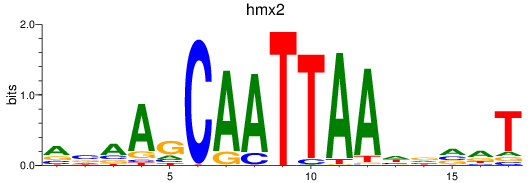
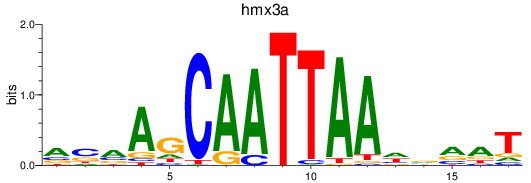
Results for hmx2_hmx3a
Z-value: 1.13
Transcription factors associated with hmx2_hmx3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx2
|
ENSDARG00000070954 | H6 family homeobox 2 |
|
hmx2
|
ENSDARG00000115364 | H6 family homeobox 2 |
|
hmx3a
|
ENSDARG00000070955 | H6 family homeobox 3a |
|
hmx3a
|
ENSDARG00000115051 | H6 family homeobox 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx3a | dr11_v1_chr17_-_21793113_21793113 | -0.35 | 5.9e-04 | Click! |
| hmx2 | dr11_v1_chr17_-_21784152_21784152 | -0.09 | 3.8e-01 | Click! |
Activity profile of hmx2_hmx3a motif
Sorted Z-values of hmx2_hmx3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38301080 | 24.24 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr25_+_31267268 | 13.93 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr5_-_25723079 | 10.68 |
ENSDART00000014013
|
gda
|
guanine deaminase |
| chr16_-_13516745 | 6.81 |
ENSDART00000145410
|
si:dkeyp-69b9.3
|
si:dkeyp-69b9.3 |
| chr6_+_29410986 | 6.65 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr8_-_39739627 | 6.01 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr24_-_10014512 | 6.00 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr11_+_36243774 | 5.82 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr24_-_9997948 | 5.73 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr8_-_39739056 | 5.69 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr13_+_24280380 | 5.69 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr1_-_19845378 | 5.63 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr10_-_1961930 | 5.45 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr21_+_40484088 | 5.39 |
ENSDART00000161919
|
coro6
|
coronin 6 |
| chr3_-_39152478 | 4.78 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr18_+_13792490 | 4.76 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr23_+_36083529 | 4.51 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr8_+_50534948 | 4.45 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr8_-_53166975 | 4.29 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr25_-_26018240 | 4.20 |
ENSDART00000150800
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr5_+_37032038 | 4.18 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr2_+_56657804 | 4.06 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr11_+_18130300 | 4.05 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr24_-_14711597 | 4.03 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr5_+_65157576 | 3.97 |
ENSDART00000159599
|
cfap157
|
cilia and flagella associated protein 157 |
| chr24_-_3477103 | 3.96 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr11_+_18157260 | 3.84 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr6_+_8315050 | 3.63 |
ENSDART00000189987
|
gcdha
|
glutaryl-CoA dehydrogenase a |
| chr5_+_24287927 | 3.57 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr25_+_20089986 | 3.49 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr20_-_23171430 | 3.47 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr14_-_33454595 | 3.36 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr20_-_40755614 | 3.32 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr21_-_35325466 | 3.31 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr9_+_23009608 | 3.22 |
ENSDART00000079879
|
si:dkey-91i10.3
|
si:dkey-91i10.3 |
| chr9_-_22821901 | 3.16 |
ENSDART00000101711
|
neb
|
nebulin |
| chr13_+_30169681 | 3.11 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr16_+_31511739 | 3.10 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr10_+_21780250 | 3.10 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr1_+_54908895 | 3.08 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr25_-_7683316 | 3.04 |
ENSDART00000128820
|
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr25_-_10503043 | 3.03 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr6_-_23931442 | 3.03 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr2_+_47623202 | 2.99 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr2_+_24304854 | 2.96 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr17_+_45737992 | 2.90 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr1_-_25966068 | 2.87 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr4_-_10599062 | 2.80 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr22_-_24757785 | 2.79 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr3_+_60721342 | 2.77 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr10_-_27046639 | 2.76 |
ENSDART00000041841
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr16_-_2818170 | 2.69 |
ENSDART00000081918
|
acot22
|
acyl-CoA thioesterase 22 |
| chr18_+_36582815 | 2.64 |
ENSDART00000059311
|
siae
|
sialic acid acetylesterase |
| chr21_+_9628854 | 2.63 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr9_-_48397702 | 2.61 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr15_-_14552101 | 2.61 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr2_+_37140448 | 2.59 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr24_+_25692802 | 2.58 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr1_-_53750522 | 2.56 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr12_+_46605600 | 2.52 |
ENSDART00000123533
|
FADS6
|
fatty acid desaturase 6 |
| chr23_+_45785563 | 2.49 |
ENSDART00000186027
|
CABZ01088036.1
|
|
| chr20_-_52846212 | 2.48 |
ENSDART00000040265
|
tbc1d7
|
TBC1 domain family, member 7 |
| chr16_-_28268201 | 2.46 |
ENSDART00000121671
ENSDART00000141911 |
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr15_-_1745408 | 2.43 |
ENSDART00000182311
|
stx1a
|
syntaxin 1A (brain) |
| chr8_+_38415374 | 2.42 |
ENSDART00000085395
|
nkx6.3
|
NK6 homeobox 3 |
| chr10_-_36591511 | 2.41 |
ENSDART00000063347
|
slc1a3b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3b |
| chr12_-_3756405 | 2.41 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr21_+_44857293 | 2.41 |
ENSDART00000134365
ENSDART00000168217 ENSDART00000065083 |
fstl4
|
follistatin-like 4 |
| chr15_+_1148074 | 2.40 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr16_+_46995398 | 2.39 |
ENSDART00000177679
|
thsd7ab
|
thrombospondin, type I, domain containing 7Ab |
| chr16_+_28881235 | 2.39 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr1_-_25966411 | 2.38 |
ENSDART00000193375
|
synpo2b
|
synaptopodin 2b |
| chr4_-_16824231 | 2.37 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr2_+_20332044 | 2.31 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr11_+_30057762 | 2.29 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr18_+_1145571 | 2.28 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr4_+_5848229 | 2.25 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr2_+_22702488 | 2.21 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr5_+_28849155 | 2.20 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr8_+_49778756 | 2.20 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr15_+_478524 | 2.19 |
ENSDART00000018062
|
si:ch1073-280e3.1
|
si:ch1073-280e3.1 |
| chr7_-_19420334 | 2.17 |
ENSDART00000173838
|
si:ch73-71d17.2
|
si:ch73-71d17.2 |
| chr11_+_37178271 | 2.16 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr3_-_50177658 | 2.14 |
ENSDART00000135309
|
zgc:114118
|
zgc:114118 |
| chr1_-_18811517 | 2.14 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr22_-_20695237 | 2.13 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr2_-_1443692 | 2.11 |
ENSDART00000004533
|
rpe65a
|
retinal pigment epithelium-specific protein 65a |
| chr23_+_45845159 | 2.11 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr5_+_28848870 | 2.11 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr17_+_15433671 | 2.11 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr6_+_11249706 | 2.10 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr10_-_42882215 | 2.08 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr21_+_39197628 | 2.07 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr18_-_8313686 | 2.06 |
ENSDART00000182187
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr11_-_4235811 | 2.03 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr7_+_58843700 | 2.03 |
ENSDART00000159500
ENSDART00000158436 |
lypla1
|
lysophospholipase I |
| chr4_+_25607101 | 2.03 |
ENSDART00000133929
|
acot14
|
acyl-CoA thioesterase 14 |
| chr2_-_47431205 | 2.00 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr23_-_27345425 | 2.00 |
ENSDART00000022042
ENSDART00000191870 |
scn8aa
|
sodium channel, voltage gated, type VIII, alpha subunit a |
| chr25_+_19106574 | 1.98 |
ENSDART00000067332
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr9_+_40939336 | 1.98 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr17_+_15433518 | 1.95 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr12_+_48395693 | 1.95 |
ENSDART00000180362
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr9_-_22822084 | 1.94 |
ENSDART00000142020
|
neb
|
nebulin |
| chr21_+_45502621 | 1.93 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr25_+_3326885 | 1.91 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr24_-_10021341 | 1.89 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr17_-_6399920 | 1.88 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr2_+_20331445 | 1.88 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr24_-_6898302 | 1.87 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr13_-_21688176 | 1.87 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr9_-_39005317 | 1.85 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr7_-_4036184 | 1.85 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr7_-_4461104 | 1.85 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr6_+_11250316 | 1.84 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr21_+_21012210 | 1.83 |
ENSDART00000190317
|
ndr1
|
nodal-related 1 |
| chr21_+_45502773 | 1.81 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr16_+_23976227 | 1.80 |
ENSDART00000193013
|
apoc4
|
apolipoprotein C-IV |
| chr17_-_6382392 | 1.80 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr1_+_29759678 | 1.80 |
ENSDART00000054059
ENSDART00000101856 |
cpb2
|
carboxypeptidase B2 (plasma) |
| chr22_-_31060579 | 1.75 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr3_+_56366395 | 1.74 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr20_-_23041275 | 1.74 |
ENSDART00000188200
|
BX511028.1
|
|
| chr12_+_24561832 | 1.74 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr23_+_21424747 | 1.73 |
ENSDART00000135440
ENSDART00000104214 |
tas1r1
|
taste receptor, type 1, member 1 |
| chr21_-_14803366 | 1.71 |
ENSDART00000190872
|
si:dkey-11o18.5
|
si:dkey-11o18.5 |
| chr24_+_5912635 | 1.70 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr19_-_45960191 | 1.69 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr11_-_23332592 | 1.67 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr2_+_9821757 | 1.67 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr3_-_35800221 | 1.67 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr13_-_29421331 | 1.67 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr16_-_52821023 | 1.67 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr12_+_16284086 | 1.66 |
ENSDART00000013360
ENSDART00000141169 |
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr17_+_16429826 | 1.65 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr15_-_21155641 | 1.65 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr3_+_17314997 | 1.63 |
ENSDART00000139763
|
stat5a
|
signal transducer and activator of transcription 5a |
| chr12_-_17897134 | 1.63 |
ENSDART00000066407
|
nptx2b
|
neuronal pentraxin IIb |
| chr10_-_40484531 | 1.63 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr21_+_6394929 | 1.62 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr6_+_11250033 | 1.61 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr16_+_22761846 | 1.61 |
ENSDART00000193028
|
CR854942.1
|
|
| chr2_+_33697340 | 1.60 |
ENSDART00000137669
|
si:dkey-31m5.3
|
si:dkey-31m5.3 |
| chr15_-_47841259 | 1.60 |
ENSDART00000155384
|
kbtbd3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr11_+_20056732 | 1.60 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr17_+_24318753 | 1.60 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr20_-_1174266 | 1.59 |
ENSDART00000023304
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr19_-_5865766 | 1.58 |
ENSDART00000191007
|
LO018585.1
|
|
| chr17_-_40397752 | 1.58 |
ENSDART00000178483
|
BX548062.1
|
|
| chr12_+_34953038 | 1.58 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr15_-_30857350 | 1.57 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr17_-_1407593 | 1.57 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr18_-_6855991 | 1.56 |
ENSDART00000135206
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr14_+_29581710 | 1.56 |
ENSDART00000188820
ENSDART00000193874 |
TENM2
|
si:dkey-34l15.2 |
| chr2_+_4894873 | 1.56 |
ENSDART00000164626
|
si:ch211-162e15.3
|
si:ch211-162e15.3 |
| chr11_+_18175893 | 1.55 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr8_-_25120231 | 1.55 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr11_+_41858807 | 1.54 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr12_+_28955766 | 1.54 |
ENSDART00000123417
ENSDART00000139347 |
znf668
|
zinc finger protein 668 |
| chr21_+_22124736 | 1.54 |
ENSDART00000130179
ENSDART00000172573 |
cul5b
|
cullin 5b |
| chr4_+_4267451 | 1.52 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr23_-_26652273 | 1.51 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr6_+_11369135 | 1.49 |
ENSDART00000151260
|
zgc:112416
|
zgc:112416 |
| chr15_+_45563656 | 1.48 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr15_-_16121496 | 1.48 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr7_-_17416092 | 1.48 |
ENSDART00000043689
ENSDART00000170003 |
nitr3a
nitr3c
|
novel immune-type receptor 3a novel immune-type receptor 3c |
| chr18_+_8937718 | 1.46 |
ENSDART00000138115
|
si:dkey-5i3.1
|
si:dkey-5i3.1 |
| chr23_-_39784368 | 1.45 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr9_-_3866707 | 1.44 |
ENSDART00000010378
|
myo3b
|
myosin IIIB |
| chr9_-_14273652 | 1.44 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr8_-_51293265 | 1.44 |
ENSDART00000127875
ENSDART00000181145 |
bmp1a
|
bone morphogenetic protein 1a |
| chr23_+_45845423 | 1.43 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr17_+_7524389 | 1.41 |
ENSDART00000067579
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr24_-_5911973 | 1.41 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr15_+_14856307 | 1.40 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr11_-_34628789 | 1.39 |
ENSDART00000192433
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr14_-_2318590 | 1.38 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr9_-_7394412 | 1.36 |
ENSDART00000145456
|
slc23a3
|
solute carrier family 23, member 3 |
| chr8_+_41035230 | 1.36 |
ENSDART00000141349
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr23_+_26946744 | 1.34 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr11_-_1509773 | 1.34 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr11_+_18183220 | 1.34 |
ENSDART00000113468
|
LO018315.10
|
|
| chr20_-_33566640 | 1.33 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr17_+_15111319 | 1.33 |
ENSDART00000185099
|
CU464134.2
|
|
| chr25_-_19433244 | 1.33 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr11_-_37691449 | 1.32 |
ENSDART00000185340
|
zgc:158258
|
zgc:158258 |
| chr10_-_43392267 | 1.32 |
ENSDART00000142872
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr20_+_48739154 | 1.31 |
ENSDART00000100262
|
zgc:110348
|
zgc:110348 |
| chr1_+_22002649 | 1.30 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr13_+_33462232 | 1.30 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr20_-_8443425 | 1.30 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr16_-_13004166 | 1.29 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr7_-_24520866 | 1.29 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr24_-_33461313 | 1.27 |
ENSDART00000163706
|
si:ch73-173p19.2
|
si:ch73-173p19.2 |
| chr23_+_33296588 | 1.27 |
ENSDART00000030368
|
si:ch211-226m16.3
|
si:ch211-226m16.3 |
| chr9_+_29548630 | 1.27 |
ENSDART00000132295
|
rnf17
|
ring finger protein 17 |
| chr6_-_21678263 | 1.27 |
ENSDART00000038777
|
si:dkey-43k4.5
|
si:dkey-43k4.5 |
| chr5_-_12407194 | 1.25 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr25_-_17587785 | 1.25 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr13_-_40316367 | 1.25 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx2_hmx3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 1.4 | 5.5 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 1.4 | 6.8 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.9 | 5.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.8 | 4.0 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.7 | 2.2 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.6 | 2.6 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.6 | 4.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.6 | 3.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.6 | 1.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.6 | 4.8 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.6 | 1.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.6 | 5.6 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.6 | 1.7 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.5 | 2.2 | GO:1901862 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.5 | 2.1 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.5 | 2.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.5 | 5.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.5 | 2.4 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.5 | 2.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.5 | 3.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 3.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.4 | 1.2 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.4 | 1.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.4 | 1.1 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.3 | 1.0 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.3 | 16.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 0.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 0.9 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.3 | 0.9 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.3 | 1.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.3 | 0.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 1.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.3 | 1.6 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.3 | 1.0 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.3 | 1.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.3 | 2.8 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.2 | 1.2 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 1.4 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.2 | 6.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 0.7 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 | 0.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 1.7 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 5.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 7.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 2.5 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 0.8 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 1.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 1.6 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 1.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 0.6 | GO:0060137 | parturition(GO:0007567) neurohypophysis development(GO:0021985) maternal process involved in parturition(GO:0060137) |
| 0.2 | 1.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 3.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 2.1 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.6 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.2 | 0.9 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.2 | 1.3 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 0.7 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.2 | 3.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.2 | 6.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 2.0 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 2.0 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 1.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.4 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.7 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.7 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.4 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.1 | 1.0 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 2.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.5 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.8 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.8 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.8 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.8 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 2.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.8 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 1.1 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.8 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.7 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 4.3 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 0.9 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 5.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 4.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 1.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 1.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.3 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.4 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.8 | GO:0090311 | regulation of protein deacetylation(GO:0090311) |
| 0.1 | 0.8 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 1.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 3.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.8 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 2.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 2.0 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 0.4 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.6 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 1.3 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 1.6 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 2.4 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.9 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.6 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 0.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 1.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.5 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 3.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 2.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 4.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 5.7 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 0.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 1.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 4.8 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 0.7 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 3.5 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 0.7 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.8 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.1 | 0.9 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 12.6 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.1 | 1.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 1.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 3.9 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.1 | 0.3 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.1 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 1.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 4.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.1 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) thymocyte migration(GO:0072679) |
| 0.0 | 0.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.7 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.7 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 1.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0048942 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.0 | 2.9 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 2.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.3 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 1.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.5 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 4.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 2.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.2 | GO:1902305 | regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 3.4 | GO:0072376 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 1.1 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.3 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.5 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.8 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.3 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 1.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.1 | GO:0071696 | ectodermal placode development(GO:0071696) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.9 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 6.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 1.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.4 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 3.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.6 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.0 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.7 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 1.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 1.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.6 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.0 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 3.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.5 | 1.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 5.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 12.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 1.2 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 2.9 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 3.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 5.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 1.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 1.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 5.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.0 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 2.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 4.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 4.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.6 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 6.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 5.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 3.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 2.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 5.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 3.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.8 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 2.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.0 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 5.2 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 2.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.9 | 3.6 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.7 | 2.2 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.7 | 4.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.6 | 2.4 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.6 | 7.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.5 | 2.1 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.5 | 2.0 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.4 | 2.6 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.4 | 2.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.4 | 2.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 2.8 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.4 | 1.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 2.9 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.3 | 1.0 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.3 | 1.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 1.0 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.3 | 0.9 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.3 | 3.9 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.3 | 0.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 0.9 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.3 | 1.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.3 | 1.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 1.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 0.8 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 4.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 2.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 2.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 5.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 1.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 1.0 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.2 | 1.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 2.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 3.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 0.9 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 1.1 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 10.7 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.2 | 1.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 0.6 | GO:0000035 | acyl binding(GO:0000035) |
| 0.2 | 1.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 0.6 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.2 | 4.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 0.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 2.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.7 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.2 | 2.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.7 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 1.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 2.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.8 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.8 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 3.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.7 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 2.1 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 3.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.4 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 1.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.7 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 1.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.4 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 1.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.1 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 7.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 4.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 2.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.6 | GO:0035254 | GKAP/Homer scaffold activity(GO:0030160) glutamate receptor binding(GO:0035254) ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.6 | GO:0015250 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.6 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.5 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 1.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 2.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 6.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 2.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.9 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.8 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 2.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.7 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 2.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 6.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 2.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 6.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 2.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 4.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.5 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 1.1 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 3.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.6 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 8.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 1.0 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 4.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.9 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 5.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 1.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 4.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.9 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.8 | 2.4 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.5 | 5.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 2.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 2.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 3.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 7.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 4.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 0.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 1.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.2 | 4.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |