Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
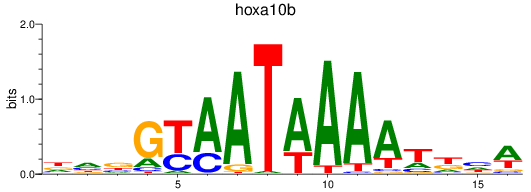
Results for hoxa10b
Z-value: 0.65
Transcription factors associated with hoxa10b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa10b
|
ENSDARG00000031337 | homeobox A10b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa10b | dr11_v1_chr16_+_20910186_20910186 | 0.19 | 7.2e-02 | Click! |
Activity profile of hoxa10b motif
Sorted Z-values of hoxa10b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_70155935 | 5.83 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr10_-_15128771 | 4.54 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr24_+_38306010 | 4.22 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr25_+_20089986 | 4.12 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr16_-_44945224 | 4.10 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr18_+_9171778 | 4.01 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr15_-_15357178 | 3.87 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr22_+_5574952 | 3.52 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr21_+_5129513 | 3.43 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr25_+_31276842 | 3.33 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr20_-_27733683 | 3.31 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr6_-_30485009 | 3.29 |
ENSDART00000025698
|
zgc:153311
|
zgc:153311 |
| chr25_+_29161609 | 2.99 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr12_-_25887864 | 2.97 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr8_+_22930627 | 2.94 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr17_+_15535501 | 2.90 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr25_+_4750972 | 2.85 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr7_+_66822229 | 2.79 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr23_-_15216654 | 2.51 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr3_-_55104310 | 2.47 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr9_+_45839260 | 2.30 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr14_-_36378494 | 2.30 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr24_+_2519761 | 2.28 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr20_-_34801181 | 2.24 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr15_-_33933790 | 2.24 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr10_+_36650222 | 2.20 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr25_+_16080181 | 2.20 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr9_+_44430974 | 2.13 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr25_-_19443421 | 2.12 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr10_-_24371312 | 2.10 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr3_+_40576447 | 1.98 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr20_-_40717900 | 1.95 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr14_-_16082806 | 1.91 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr8_-_36140405 | 1.88 |
ENSDART00000182806
|
CT583723.2
|
|
| chr25_+_34984333 | 1.87 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr16_+_35916371 | 1.87 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr11_-_21031773 | 1.86 |
ENSDART00000065985
|
fmoda
|
fibromodulin a |
| chr7_+_51103232 | 1.82 |
ENSDART00000073827
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr16_+_36126310 | 1.80 |
ENSDART00000166040
ENSDART00000189802 |
sh3bp5b
|
SH3-domain binding protein 5b (BTK-associated) |
| chr6_+_35362225 | 1.79 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr12_-_5728755 | 1.75 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr12_+_23912074 | 1.72 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr12_+_11352630 | 1.69 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr3_+_29714775 | 1.68 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr15_+_43166511 | 1.66 |
ENSDART00000011737
|
flj13639
|
flj13639 |
| chr24_-_21512425 | 1.64 |
ENSDART00000184857
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr8_-_17064243 | 1.63 |
ENSDART00000185313
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr9_-_40765868 | 1.63 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr8_-_39952727 | 1.63 |
ENSDART00000181310
|
cabp1a
|
calcium binding protein 1a |
| chr5_-_31856681 | 1.63 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr7_+_1473929 | 1.62 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr21_-_2299002 | 1.60 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr24_-_25256230 | 1.56 |
ENSDART00000155780
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr1_-_26131088 | 1.53 |
ENSDART00000193973
ENSDART00000054209 |
cdkn2a/b
|
cyclin-dependent kinase inhibitor 2A/B (p15, inhibits CDK4) |
| chr12_+_10053852 | 1.52 |
ENSDART00000078497
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr9_-_31278048 | 1.51 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr9_+_44430705 | 1.49 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_-_31661609 | 1.45 |
ENSDART00000144383
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr24_-_15159658 | 1.42 |
ENSDART00000142473
|
rttn
|
rotatin |
| chr1_+_46194333 | 1.42 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr12_+_35650321 | 1.42 |
ENSDART00000190446
|
BX255898.1
|
|
| chr20_-_25709247 | 1.42 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr22_-_881725 | 1.40 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr10_+_18877362 | 1.40 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr24_+_23791758 | 1.38 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr15_-_1844048 | 1.36 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr8_-_11324143 | 1.35 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr7_-_44672095 | 1.32 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr7_+_51103416 | 1.31 |
ENSDART00000174236
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr12_+_27285994 | 1.30 |
ENSDART00000087204
|
dusp3a
|
dual specificity phosphatase 3a |
| chr20_+_37393134 | 1.30 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr2_-_31664615 | 1.28 |
ENSDART00000190546
ENSDART00000111810 |
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr20_-_23219964 | 1.24 |
ENSDART00000144933
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr21_-_26691959 | 1.21 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr9_-_32604414 | 1.21 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr1_-_22512063 | 1.20 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr14_+_34966598 | 1.20 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr19_-_20113696 | 1.20 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr11_+_42730639 | 1.19 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr9_-_16853462 | 1.18 |
ENSDART00000160273
|
CT573248.2
|
|
| chr5_-_31857345 | 1.17 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr9_+_34232503 | 1.17 |
ENSDART00000132836
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr3_+_26135502 | 1.17 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr21_-_40173821 | 1.16 |
ENSDART00000180667
|
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr17_+_43595692 | 1.16 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr10_-_34889053 | 1.14 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr15_+_16387088 | 1.13 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr20_-_29499363 | 1.13 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr13_+_25455319 | 1.12 |
ENSDART00000145948
|
pkd2l1
|
polycystic kidney disease 2-like 1 |
| chr23_+_28092083 | 1.10 |
ENSDART00000053958
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr14_+_36226243 | 1.09 |
ENSDART00000052569
|
pitx2
|
paired-like homeodomain 2 |
| chr19_-_47571797 | 1.09 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr15_+_3825117 | 1.08 |
ENSDART00000183315
|
CABZ01061591.1
|
|
| chr11_+_29856032 | 1.08 |
ENSDART00000079150
|
grpr
|
gastrin-releasing peptide receptor |
| chr18_+_6857071 | 1.08 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr25_+_3549841 | 1.07 |
ENSDART00000164030
|
ccdc77
|
coiled-coil domain containing 77 |
| chr6_-_6448519 | 1.06 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr8_+_3820134 | 1.06 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr16_-_24561354 | 1.03 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr3_+_24275597 | 1.03 |
ENSDART00000183926
ENSDART00000169765 |
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr19_-_47571456 | 1.03 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr1_+_57371447 | 1.01 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr23_-_12014931 | 1.01 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr6_+_11989537 | 1.00 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr6_-_40697585 | 1.00 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr10_-_25816558 | 0.99 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr23_-_21215520 | 0.98 |
ENSDART00000143206
|
megf6a
|
multiple EGF-like-domains 6a |
| chr22_+_9806867 | 0.97 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr4_+_35594200 | 0.97 |
ENSDART00000193902
|
si:dkeyp-4c4.2
|
si:dkeyp-4c4.2 |
| chr16_+_21789703 | 0.95 |
ENSDART00000153617
|
trim108
|
tripartite motif containing 108 |
| chr19_-_31402429 | 0.95 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr2_+_35733335 | 0.95 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr1_-_55873178 | 0.94 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr23_-_21215311 | 0.94 |
ENSDART00000112424
|
megf6a
|
multiple EGF-like-domains 6a |
| chr21_-_15929041 | 0.94 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr1_+_52392511 | 0.93 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr2_+_19163965 | 0.93 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr25_+_3549401 | 0.92 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr1_-_55068941 | 0.92 |
ENSDART00000152143
ENSDART00000152590 |
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr21_+_6556635 | 0.91 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr10_-_34772211 | 0.89 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr24_-_3407507 | 0.88 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr25_-_14433503 | 0.88 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr22_+_15959844 | 0.87 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr2_+_6126086 | 0.86 |
ENSDART00000179962
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr15_-_19772372 | 0.85 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr7_+_34290051 | 0.83 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr16_-_32649929 | 0.83 |
ENSDART00000136161
|
faxcb
|
failed axon connections homolog b |
| chr10_-_31104983 | 0.82 |
ENSDART00000186941
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr3_-_21037840 | 0.82 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr25_+_7423770 | 0.82 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr15_-_1843831 | 0.81 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_-_36591511 | 0.81 |
ENSDART00000063347
|
slc1a3b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3b |
| chr11_+_21053488 | 0.81 |
ENSDART00000189860
|
zgc:113307
|
zgc:113307 |
| chr23_+_45538932 | 0.81 |
ENSDART00000135602
|
si:ch73-290k24.6
|
si:ch73-290k24.6 |
| chr2_+_7715810 | 0.80 |
ENSDART00000130781
|
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr12_+_10599604 | 0.80 |
ENSDART00000152538
|
lrrc3ca
|
leucine rich repeat containing 3Ca |
| chr14_-_45604667 | 0.80 |
ENSDART00000167016
|
si:ch211-276i12.9
|
si:ch211-276i12.9 |
| chr21_-_23475361 | 0.79 |
ENSDART00000156658
ENSDART00000157454 |
ncam1a
|
neural cell adhesion molecule 1a |
| chr16_+_31942527 | 0.78 |
ENSDART00000188334
ENSDART00000188643 ENSDART00000058496 |
phc1
|
polyhomeotic homolog 1 |
| chr20_-_2641233 | 0.78 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr24_+_19578935 | 0.78 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr1_+_2321384 | 0.77 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr20_-_9273669 | 0.77 |
ENSDART00000175069
|
syt14b
|
synaptotagmin XIVb |
| chr6_+_13933464 | 0.76 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr20_-_36416922 | 0.74 |
ENSDART00000019145
|
lbr
|
lamin B receptor |
| chr8_+_23105117 | 0.73 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr21_+_39423974 | 0.72 |
ENSDART00000031470
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr3_+_33761549 | 0.72 |
ENSDART00000169337
|
ier2a
|
immediate early response 2a |
| chr10_-_11840353 | 0.71 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr23_+_41799748 | 0.70 |
ENSDART00000144257
|
pdyn
|
prodynorphin |
| chr3_-_29910547 | 0.70 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr19_+_7424347 | 0.70 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr12_+_40905427 | 0.69 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr18_-_39288894 | 0.68 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr5_-_57723929 | 0.67 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr12_+_13205955 | 0.67 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr14_+_15597049 | 0.66 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr24_-_23784701 | 0.66 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr22_-_12337781 | 0.66 |
ENSDART00000188357
ENSDART00000123574 |
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr23_-_24253200 | 0.66 |
ENSDART00000114840
|
zbtb17
|
zinc finger and BTB domain containing 17 |
| chr16_-_32671998 | 0.66 |
ENSDART00000161395
|
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr4_+_17336557 | 0.66 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr8_-_23573084 | 0.65 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr4_-_39171837 | 0.64 |
ENSDART00000179734
|
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr25_-_4733221 | 0.63 |
ENSDART00000172689
|
drd4a
|
dopamine receptor D4a |
| chr3_-_58650057 | 0.63 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr19_+_636886 | 0.63 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr5_-_24997293 | 0.63 |
ENSDART00000177478
|
emid1
|
EMI domain containing 1 |
| chr23_+_45200481 | 0.62 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr2_-_16380283 | 0.62 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr2_+_58008980 | 0.61 |
ENSDART00000171264
|
si:ch211-155e24.3
|
si:ch211-155e24.3 |
| chr10_-_29768556 | 0.60 |
ENSDART00000052787
|
vps11
|
vacuolar protein sorting 11 |
| chr13_-_8892514 | 0.59 |
ENSDART00000144553
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr15_+_42285643 | 0.59 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr23_-_29505645 | 0.59 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr2_+_2223837 | 0.58 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr25_+_15933411 | 0.58 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr17_-_22010668 | 0.57 |
ENSDART00000031998
|
slc22a7b.1
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 1 |
| chr5_+_20453874 | 0.57 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr13_-_7575216 | 0.56 |
ENSDART00000159443
|
pitx3
|
paired-like homeodomain 3 |
| chr5_+_24063046 | 0.56 |
ENSDART00000051548
ENSDART00000133355 ENSDART00000142268 |
gps2
|
G protein pathway suppressor 2 |
| chr11_-_5953636 | 0.55 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr2_-_30912922 | 0.55 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr21_-_26071773 | 0.55 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr10_-_43771447 | 0.54 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr6_-_10728921 | 0.53 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr22_-_31060579 | 0.50 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr15_-_47857687 | 0.50 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr16_-_32671782 | 0.49 |
ENSDART00000123980
|
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr21_-_38853737 | 0.49 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr14_+_10625112 | 0.49 |
ENSDART00000143377
ENSDART00000136480 |
nup62l
|
nucleoporin 62 like |
| chr21_-_14878220 | 0.48 |
ENSDART00000131237
|
ulk1b
|
unc-51 like autophagy activating kinase 1 |
| chr23_+_12455295 | 0.48 |
ENSDART00000143752
ENSDART00000135785 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr18_-_50614024 | 0.47 |
ENSDART00000171207
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr17_-_30652738 | 0.47 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr8_-_31416403 | 0.47 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr1_-_12394048 | 0.47 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr5_+_54555567 | 0.46 |
ENSDART00000171159
|
anapc2
|
anaphase promoting complex subunit 2 |
| chr5_+_58492699 | 0.46 |
ENSDART00000181584
|
FQ378016.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa10b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:2000051 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.9 | 3.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.8 | 4.0 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.6 | 3.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.6 | 1.7 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.6 | 3.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 1.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.4 | 1.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.4 | 2.0 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.3 | 1.0 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.3 | 1.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 1.2 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.3 | 3.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 1.2 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.3 | 2.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 1.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.3 | 2.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 0.8 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.2 | 2.9 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.2 | 1.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 3.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 1.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 1.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 0.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 1.6 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.2 | 0.6 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 1.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 7.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.8 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 1.9 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 1.4 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.8 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 1.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 2.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.7 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 1.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.4 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 1.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 3.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 1.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.7 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 4.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.1 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.1 | 4.5 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 0.9 | GO:0030809 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.1 | 1.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 3.1 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 3.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 2.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.8 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.9 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.7 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.3 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 1.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.6 | GO:0070306 | adenohypophysis development(GO:0021984) lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.8 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 3.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.6 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 1.2 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 2.2 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 1.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.3 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.7 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 2.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 0.7 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.2 | 2.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 7.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.5 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.9 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 2.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 3.2 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 4.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 2.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 2.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.8 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 3.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 1.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 2.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 3.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 3.2 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 2.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 2.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 1.1 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 4.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.2 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 0.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.2 | 3.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 3.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 0.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 3.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 2.5 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 2.8 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.3 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 1.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 3.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.8 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 1.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.2 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 2.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 1.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 2.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.6 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 4.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 4.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |