Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
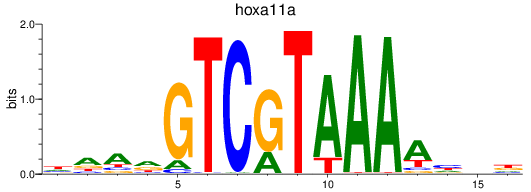
Results for hoxa11a
Z-value: 0.30
Transcription factors associated with hoxa11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11a
|
ENSDARG00000104162 | homeobox A11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11a | dr11_v1_chr19_+_19737214_19737300 | 0.14 | 1.9e-01 | Click! |
Activity profile of hoxa11a motif
Sorted Z-values of hoxa11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_27868183 | 2.34 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr10_+_2587234 | 1.60 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr7_+_20524064 | 1.40 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr2_-_13254821 | 1.39 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr17_-_23727978 | 1.38 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr7_-_41014773 | 1.31 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr14_-_48348973 | 1.26 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr6_+_42338309 | 1.21 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr2_-_10338759 | 1.16 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr15_+_9297340 | 1.13 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr3_+_24190207 | 1.08 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr7_+_25036188 | 1.07 |
ENSDART00000163957
ENSDART00000169749 |
sb:cb1058
|
sb:cb1058 |
| chr24_-_40009446 | 1.04 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr8_-_14049404 | 1.02 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr2_-_13254594 | 0.99 |
ENSDART00000155671
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr15_+_8767650 | 0.98 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr9_-_3671911 | 0.89 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr7_+_9308625 | 0.87 |
ENSDART00000084598
|
selenos
|
selenoprotein S |
| chr13_-_25196758 | 0.83 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr6_+_49551614 | 0.80 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr15_+_36115955 | 0.79 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr18_-_2433011 | 0.79 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr20_-_44576949 | 0.78 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr1_+_36722122 | 0.73 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr13_-_9875538 | 0.59 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr6_-_53334259 | 0.59 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr5_-_35301800 | 0.54 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr17_+_44463230 | 0.51 |
ENSDART00000130311
|
naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr6_-_40744720 | 0.51 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr11_-_36051004 | 0.47 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr12_-_26851726 | 0.44 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr16_-_6205790 | 0.44 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr20_+_37820992 | 0.42 |
ENSDART00000064692
|
tatdn3
|
TatD DNase domain containing 3 |
| chr15_+_44093286 | 0.41 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr3_+_36284986 | 0.40 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr23_-_33750135 | 0.39 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr23_-_33750307 | 0.38 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr17_-_31695217 | 0.36 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr9_-_29039506 | 0.35 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr4_+_39368978 | 0.33 |
ENSDART00000160640
|
si:dkey-261o4.1
|
si:dkey-261o4.1 |
| chr2_-_37874647 | 0.33 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr4_-_20108833 | 0.31 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr5_-_32489796 | 0.30 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr22_-_33916620 | 0.29 |
ENSDART00000191276
|
CR974456.1
|
|
| chr4_-_9586713 | 0.29 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr14_-_31060082 | 0.29 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr25_-_18002498 | 0.28 |
ENSDART00000158688
|
cep290
|
centrosomal protein 290 |
| chr24_-_31223232 | 0.28 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr6_+_33885828 | 0.26 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr2_-_43851915 | 0.26 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr11_+_41135055 | 0.26 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr2_+_43851983 | 0.25 |
ENSDART00000126413
|
nlrb5
|
NOD-like receptor family B, member 5 |
| chr25_-_19395156 | 0.24 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr5_+_36611128 | 0.24 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr5_+_59397739 | 0.24 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr2_-_9259283 | 0.24 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr25_-_18002937 | 0.23 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr22_+_2239974 | 0.23 |
ENSDART00000141993
|
znf1144
|
zinc finger protein 1144 |
| chr9_+_20483846 | 0.21 |
ENSDART00000192067
ENSDART00000145111 |
parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr20_-_46541834 | 0.20 |
ENSDART00000060685
ENSDART00000181720 |
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr24_+_20658760 | 0.20 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr2_-_43852207 | 0.20 |
ENSDART00000192627
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr25_+_3788443 | 0.19 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr15_-_734203 | 0.19 |
ENSDART00000177867
ENSDART00000154918 |
si:dkey-7i4.21
BX004964.1
|
si:dkey-7i4.21 |
| chr24_+_20658942 | 0.18 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr20_+_34671386 | 0.17 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr1_-_56213723 | 0.16 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr11_+_25560632 | 0.15 |
ENSDART00000033914
|
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr11_+_25560072 | 0.14 |
ENSDART00000124131
ENSDART00000147179 |
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr16_-_24561354 | 0.13 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr12_-_11560794 | 0.10 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr7_-_30624435 | 0.09 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr2_+_54696042 | 0.08 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr10_-_25561751 | 0.08 |
ENSDART00000147089
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr21_+_13127742 | 0.07 |
ENSDART00000179221
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr15_-_14552101 | 0.07 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr21_-_25618175 | 0.06 |
ENSDART00000133512
|
fosl1b
|
FOS-like antigen 1b |
| chr20_+_43379029 | 0.06 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr2_-_6065416 | 0.06 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr11_-_21303946 | 0.03 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr21_+_11865972 | 0.03 |
ENSDART00000081676
|
ubap1
|
ubiquitin associated protein 1 |
| chr16_-_47426482 | 0.02 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr3_-_18737126 | 0.01 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr19_-_22507715 | 0.01 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr12_-_34713690 | 0.01 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.5 | 2.3 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.4 | 1.3 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.3 | 1.7 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 1.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 0.9 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 0.9 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.4 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.8 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 1.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 1.0 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.6 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.2 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.5 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.9 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 0.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.4 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 1.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.8 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 2.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 1.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 2.4 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |