Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
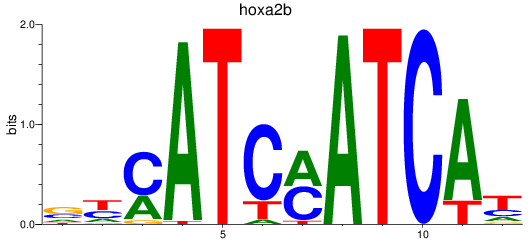
Results for hoxa2b
Z-value: 0.78
Transcription factors associated with hoxa2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa2b
|
ENSDARG00000023031 | homeobox A2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa2b | dr11_v1_chr16_+_20926673_20926673 | 0.07 | 5.2e-01 | Click! |
Activity profile of hoxa2b motif
Sorted Z-values of hoxa2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_38301080 | 56.81 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr11_+_36243774 | 22.15 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr12_-_11457625 | 9.17 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr3_-_32818607 | 7.26 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr1_+_17676745 | 5.73 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr7_+_29952719 | 5.68 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr9_-_45601103 | 5.32 |
ENSDART00000180465
|
agr1
|
anterior gradient 1 |
| chr8_+_21437908 | 5.31 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr6_+_13201358 | 5.18 |
ENSDART00000190290
|
CT009620.1
|
|
| chr14_+_15495088 | 5.02 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr23_+_44611864 | 4.78 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr20_-_53992952 | 4.57 |
ENSDART00000170138
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr14_+_15331486 | 4.52 |
ENSDART00000172077
ENSDART00000183370 ENSDART00000182467 |
SPINK2
si:dkey-203a12.5
|
si:dkey-203a12.4 si:dkey-203a12.5 |
| chr10_-_20445549 | 4.40 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr4_+_7391110 | 4.28 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr5_+_32221755 | 4.27 |
ENSDART00000125917
|
myhc4
|
myosin heavy chain 4 |
| chr13_+_22480496 | 4.06 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr14_+_15543331 | 3.94 |
ENSDART00000167025
|
si:dkey-203a12.7
|
si:dkey-203a12.7 |
| chr12_-_26430507 | 3.92 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr5_+_61976511 | 3.88 |
ENSDART00000050885
|
ehd2a
|
EH-domain containing 2a |
| chr14_+_15231097 | 3.85 |
ENSDART00000172430
|
si:dkey-203a12.3
|
si:dkey-203a12.3 |
| chr25_+_20216159 | 3.85 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr25_+_20215964 | 3.78 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr12_-_4346085 | 3.77 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr7_-_4461104 | 3.71 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr7_+_4386753 | 3.69 |
ENSDART00000172915
|
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr6_+_3640381 | 3.63 |
ENSDART00000172078
|
col28a2b
|
collagen, type XXVIII, alpha 2b |
| chr11_+_29671661 | 3.56 |
ENSDART00000024318
ENSDART00000165024 |
rnf207a
|
ring finger protein 207a |
| chr13_+_22249636 | 3.22 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr14_+_15257658 | 3.15 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr25_+_35020529 | 3.06 |
ENSDART00000158016
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr4_+_40427249 | 2.96 |
ENSDART00000151927
|
si:ch211-218h8.3
|
si:ch211-218h8.3 |
| chr12_+_48241841 | 2.86 |
ENSDART00000168616
|
ppa1a
|
pyrophosphatase (inorganic) 1a |
| chr9_-_712308 | 2.85 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr23_-_18609475 | 2.84 |
ENSDART00000104524
|
zgc:158296
|
zgc:158296 |
| chr24_+_5789790 | 2.80 |
ENSDART00000189600
|
BX470065.1
|
|
| chr3_+_34683096 | 2.79 |
ENSDART00000084432
|
dusp3b
|
dual specificity phosphatase 3b |
| chr14_+_15430991 | 2.75 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr23_+_36095260 | 2.74 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr24_+_5789582 | 2.73 |
ENSDART00000141504
|
BX470065.1
|
|
| chr7_-_17337233 | 2.73 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr11_+_21096339 | 2.70 |
ENSDART00000124574
|
il19l
|
interleukin 19 like |
| chr3_+_33300522 | 2.69 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr25_-_35497055 | 2.66 |
ENSDART00000009271
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr25_-_31396479 | 2.64 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr5_-_38820046 | 2.64 |
ENSDART00000182886
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr7_-_7810348 | 2.62 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr4_-_9909371 | 2.59 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr20_-_29420713 | 2.57 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr7_-_41858513 | 2.51 |
ENSDART00000109918
|
mylk3
|
myosin light chain kinase 3 |
| chr3_-_26109322 | 2.49 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr13_+_9432501 | 2.42 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr7_-_20453661 | 2.38 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr2_-_722156 | 2.36 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr23_+_20408227 | 2.35 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr1_-_9227804 | 2.34 |
ENSDART00000190360
|
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr21_+_11969603 | 2.32 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr1_+_15216988 | 2.31 |
ENSDART00000189206
ENSDART00000181639 |
itln1
|
intelectin 1 |
| chr2_+_17451656 | 2.30 |
ENSDART00000163620
|
BX640408.1
|
|
| chr7_+_1473929 | 2.30 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr3_-_20957011 | 2.28 |
ENSDART00000159787
ENSDART00000180531 |
ndufa4l
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4, like |
| chr4_-_14915268 | 2.27 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr1_-_25966068 | 2.27 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr4_-_43731342 | 2.26 |
ENSDART00000146627
|
si:ch211-226o13.3
|
si:ch211-226o13.3 |
| chr6_-_49510553 | 2.25 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr6_-_13206255 | 2.19 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr19_-_9648542 | 2.19 |
ENSDART00000172628
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr1_-_9228007 | 2.18 |
ENSDART00000147277
ENSDART00000135219 |
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr7_+_74150839 | 2.17 |
ENSDART00000160195
|
ppp1cbl
|
protein phosphatase 1, catalytic subunit, beta isoform, like |
| chr7_+_35229645 | 2.15 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr13_+_7442023 | 2.11 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr10_+_35417099 | 2.10 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr7_-_5162292 | 2.08 |
ENSDART00000084218
|
zgc:195075
|
zgc:195075 |
| chr3_-_34095221 | 2.07 |
ENSDART00000164235
ENSDART00000151377 |
ighv1-4
ighv5-4
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 5-4 |
| chr8_-_52745141 | 2.06 |
ENSDART00000168359
ENSDART00000168252 |
fgf17
|
fibroblast growth factor 17 |
| chr1_+_41454210 | 2.05 |
ENSDART00000148251
|
si:ch211-89o9.4
|
si:ch211-89o9.4 |
| chr17_-_49800869 | 2.05 |
ENSDART00000156264
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr12_-_26406323 | 2.02 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr3_-_32596394 | 1.99 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr4_+_76575585 | 1.98 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr19_+_28256076 | 1.96 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr21_-_32684570 | 1.92 |
ENSDART00000162873
|
adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr4_-_44500201 | 1.92 |
ENSDART00000150460
|
si:dkeyp-100h4.7
|
si:dkeyp-100h4.7 |
| chr10_+_439692 | 1.91 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr21_-_22737228 | 1.89 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr19_+_24575077 | 1.89 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr3_+_20001608 | 1.88 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr7_-_16562200 | 1.88 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr25_-_29072162 | 1.87 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr4_-_42408339 | 1.85 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr1_-_10914523 | 1.84 |
ENSDART00000007013
|
dmd
|
dystrophin |
| chr22_+_1123873 | 1.84 |
ENSDART00000137708
|
si:ch1073-181h11.2
|
si:ch1073-181h11.2 |
| chr24_+_13735616 | 1.83 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr4_-_16412084 | 1.83 |
ENSDART00000188460
|
dcn
|
decorin |
| chr7_-_15370042 | 1.82 |
ENSDART00000128183
|
ANPEP
|
si:ch211-276a23.5 |
| chr8_-_42624454 | 1.78 |
ENSDART00000075550
|
kazald3
|
Kazal-type serine peptidase inhibitor domain 3 |
| chr24_+_38201089 | 1.77 |
ENSDART00000132338
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr6_+_50451337 | 1.76 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr3_-_31254979 | 1.73 |
ENSDART00000130280
|
apnl
|
actinoporin-like protein |
| chr25_+_11456696 | 1.73 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr7_+_65533307 | 1.73 |
ENSDART00000156301
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr6_-_39037613 | 1.72 |
ENSDART00000098906
|
tns2b
|
tensin 2b |
| chr1_-_681116 | 1.70 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr17_+_27434626 | 1.69 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr13_-_1349922 | 1.69 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr24_-_29822913 | 1.67 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr3_+_27027781 | 1.67 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr1_+_57145072 | 1.67 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr4_+_44454270 | 1.67 |
ENSDART00000150482
|
si:dkeyp-100h4.4
|
si:dkeyp-100h4.4 |
| chr3_-_34032019 | 1.65 |
ENSDART00000151779
|
ighv13-2
|
immunoglobulin heavy variable 13-2 |
| chr10_+_2807136 | 1.65 |
ENSDART00000126440
ENSDART00000131435 |
aebp1
|
AE binding protein 1 |
| chr24_+_38209946 | 1.64 |
ENSDART00000058204
|
CU896602.2
|
|
| chr24_-_40744672 | 1.64 |
ENSDART00000160672
|
CU633479.1
|
|
| chr1_-_59095035 | 1.63 |
ENSDART00000130791
|
MFAP4 (1 of many)
|
wu:fj11g02 |
| chr6_+_149405 | 1.61 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr1_+_55608520 | 1.61 |
ENSDART00000152307
|
adgre18
|
adhesion G protein-coupled receptor E18 |
| chr22_-_24297510 | 1.60 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr15_-_43625549 | 1.59 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr13_-_39736938 | 1.58 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr16_+_20904754 | 1.58 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr2_+_23731194 | 1.58 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr8_+_27807974 | 1.57 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr4_-_77331797 | 1.57 |
ENSDART00000162325
|
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr5_-_7199998 | 1.56 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr1_+_58370526 | 1.55 |
ENSDART00000067775
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr14_-_6666854 | 1.55 |
ENSDART00000133031
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr7_+_35229805 | 1.55 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr16_+_1383914 | 1.54 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr16_-_35952789 | 1.53 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr21_+_18997511 | 1.52 |
ENSDART00000145591
|
rpl17
|
ribosomal protein L17 |
| chr14_+_15191176 | 1.52 |
ENSDART00000183447
ENSDART00000193093 ENSDART00000169309 |
si:dkey-203a12.2
|
si:dkey-203a12.2 |
| chr12_-_4475890 | 1.52 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr7_+_20503344 | 1.52 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr3_-_1434135 | 1.51 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr3_+_23768898 | 1.51 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr9_+_48007081 | 1.50 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr23_+_32101361 | 1.50 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr8_-_14091886 | 1.49 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr16_-_13388821 | 1.48 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr21_-_30714665 | 1.47 |
ENSDART00000128011
|
tnfsf10l3
|
tumor necrosis factor (ligand) superfamily, member 10 like 3 |
| chr4_+_77948970 | 1.46 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_-_61116258 | 1.45 |
ENSDART00000009194
ENSDART00000156978 |
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr23_+_4689626 | 1.45 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr3_+_23752150 | 1.45 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr5_+_45007962 | 1.43 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr21_-_41025340 | 1.41 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr24_-_26310854 | 1.41 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr21_-_40174647 | 1.41 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr23_-_7674902 | 1.40 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr20_+_34455645 | 1.38 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr12_+_6195191 | 1.38 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr15_-_12270857 | 1.36 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr17_-_45125537 | 1.35 |
ENSDART00000113552
|
zgc:163014
|
zgc:163014 |
| chr23_-_19500559 | 1.35 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr4_-_77332032 | 1.34 |
ENSDART00000168628
ENSDART00000172025 |
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr17_+_6563307 | 1.33 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr3_-_43872889 | 1.33 |
ENSDART00000170553
|
mslna
|
mesothelin a |
| chr21_-_308852 | 1.32 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr3_+_58655375 | 1.32 |
ENSDART00000186908
|
FP016018.4
|
|
| chr12_+_48634927 | 1.32 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr25_-_29611476 | 1.32 |
ENSDART00000154458
|
si:ch211-253p14.2
|
si:ch211-253p14.2 |
| chr16_+_40954481 | 1.31 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr7_+_4911404 | 1.30 |
ENSDART00000137385
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr4_+_20576857 | 1.29 |
ENSDART00000125340
|
BX248410.2
|
|
| chr1_-_43727418 | 1.29 |
ENSDART00000133715
ENSDART00000074597 ENSDART00000132542 ENSDART00000181792 |
bdh2
SLC9B2
|
3-hydroxybutyrate dehydrogenase, type 2 si:dkey-162b23.4 |
| chr17_-_49978986 | 1.29 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr7_+_2228276 | 1.29 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr13_-_42757565 | 1.29 |
ENSDART00000161950
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr1_+_51496862 | 1.29 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr5_-_23840920 | 1.28 |
ENSDART00000143279
|
GBGT1 (1 of many)
|
si:ch211-135f11.5 |
| chr22_+_18477934 | 1.28 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr1_-_40735381 | 1.28 |
ENSDART00000093269
|
zgc:153642
|
zgc:153642 |
| chr19_-_42607451 | 1.28 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr3_+_1749793 | 1.28 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr7_+_34297271 | 1.28 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_-_15641686 | 1.28 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr2_+_47935476 | 1.28 |
ENSDART00000140555
|
ftr26
|
finTRIM family, member 26 |
| chr2_-_32457919 | 1.27 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr7_-_25697285 | 1.26 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr7_+_53156810 | 1.26 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr3_+_57820913 | 1.26 |
ENSDART00000168101
|
CU571328.1
|
|
| chr9_-_9842149 | 1.25 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr10_-_22127942 | 1.25 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr24_-_2381143 | 1.24 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr24_+_14859238 | 1.23 |
ENSDART00000113859
ENSDART00000138227 |
crispld1a
|
cysteine-rich secretory protein LCCL domain containing 1a |
| chr15_+_17251191 | 1.22 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr8_+_27807266 | 1.21 |
ENSDART00000170037
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr25_+_24616717 | 1.21 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
| chr7_-_51793333 | 1.21 |
ENSDART00000180654
|
BX957362.5
|
|
| chr20_-_54924593 | 1.20 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
| chr9_+_22351443 | 1.19 |
ENSDART00000080054
|
crygs3
|
crystallin, gamma S3 |
| chr5_+_9259971 | 1.18 |
ENSDART00000163060
|
susd1
|
sushi domain containing 1 |
| chr24_-_14209202 | 1.17 |
ENSDART00000180674
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr15_+_32268790 | 1.17 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr23_+_43177290 | 1.17 |
ENSDART00000193300
ENSDART00000186065 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr7_+_4922104 | 1.17 |
ENSDART00000135154
|
si:dkey-28d5.11
|
si:dkey-28d5.11 |
| chr17_-_50301313 | 1.16 |
ENSDART00000125772
|
si:ch73-50f9.4
|
si:ch73-50f9.4 |
| chr11_-_25538341 | 1.16 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr25_-_16742636 | 1.15 |
ENSDART00000192486
ENSDART00000192711 |
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr19_+_32553874 | 1.15 |
ENSDART00000078197
|
heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 26.5 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 1.4 | 5.7 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 1.1 | 4.6 | GO:1904407 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 1.1 | 4.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.8 | 3.2 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.6 | 4.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.6 | 1.7 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.5 | 1.9 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.5 | 0.9 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.5 | 1.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.5 | 5.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.5 | 1.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.4 | 1.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 1.7 | GO:0042117 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.4 | 2.5 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 | 1.6 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 3.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.4 | 1.1 | GO:0071498 | cellular response to osmotic stress(GO:0071470) cellular response to fluid shear stress(GO:0071498) |
| 0.4 | 1.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 | 1.8 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.3 | 2.1 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.3 | 1.4 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.3 | 9.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.3 | 1.0 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.3 | 1.0 | GO:0070228 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.3 | 1.3 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.3 | 1.6 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 2.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.5 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.3 | 1.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 | 1.7 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.3 | 7.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 1.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 0.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 2.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 1.1 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 0.7 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 1.6 | GO:0086011 | membrane repolarization during action potential(GO:0086011) cardiac muscle cell membrane repolarization(GO:0099622) |
| 0.2 | 0.9 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.2 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 11.0 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.2 | 1.3 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 1.5 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.2 | 2.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 0.6 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.2 | 65.1 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.2 | 0.8 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 1.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 2.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.2 | 0.9 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 1.3 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.2 | 3.3 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.2 | 0.3 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.2 | 0.9 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.7 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.2 | 0.6 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 2.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0002513 | tolerance induction(GO:0002507) tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.8 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.6 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 2.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 1.3 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 4.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.4 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.1 | 0.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 1.1 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.1 | 0.7 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 1.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.3 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 0.8 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 2.6 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 0.7 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 1.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.5 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.5 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.4 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.4 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 3.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.4 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 3.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.2 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.9 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.1 | 0.5 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 1.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.6 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.4 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 4.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.6 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 1.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 3.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.4 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.7 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.1 | 0.9 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 5.0 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 3.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.1 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.1 | 1.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.5 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 3.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.4 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.8 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.5 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.2 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 5.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.5 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.3 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.6 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.8 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 1.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 3.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.3 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.2 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 1.5 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.4 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 5.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.5 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 8.4 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.6 | GO:0051209 | sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 3.0 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 1.2 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.0 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.5 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.5 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.2 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 3.8 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 1.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.3 | GO:0045089 | positive regulation of defense response(GO:0031349) positive regulation of innate immune response(GO:0045089) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.2 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.1 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.5 | GO:0051607 | defense response to virus(GO:0051607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.7 | 28.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.6 | 4.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 1.7 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 2.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 1.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 14.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 2.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 4.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.2 | 0.6 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 1.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 18.4 | GO:0031674 | I band(GO:0031674) |
| 0.2 | 0.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 1.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 13.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 6.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 7.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 3.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 5.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 9.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.1 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 2.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 3.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 17.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 3.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.9 | 8.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.7 | 2.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.7 | 7.6 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.7 | 2.7 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.6 | 4.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 2.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 1.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 1.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.5 | 1.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.5 | 1.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.4 | 1.7 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.4 | 1.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 11.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 1.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 1.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 2.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.3 | 0.9 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.3 | 1.5 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.3 | 1.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.3 | 0.8 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.3 | 4.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 2.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 2.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 1.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.9 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.2 | 5.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 2.0 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 0.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 2.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.9 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 4.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.9 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 2.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 3.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.0 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.9 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 22.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 1.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 1.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.7 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.7 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 1.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.5 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.3 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.5 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.2 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 1.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 3.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 3.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 3.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.6 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.6 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 4.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.6 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 8.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 4.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 2.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 4.2 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.0 | GO:0031697 | adrenergic receptor binding(GO:0031690) beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.6 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 4.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 1.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.0 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 6.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 2.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 1.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 1.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 1.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 3.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 5.6 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 4.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.9 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 3.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.7 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |