Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
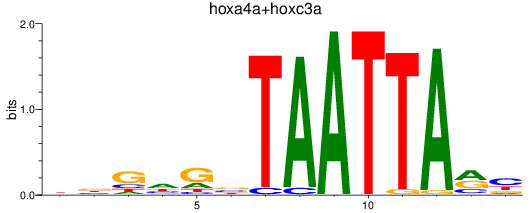
Results for hoxa4a+hoxc3a
Z-value: 0.67
Transcription factors associated with hoxa4a+hoxc3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc3a
|
ENSDARG00000070339 | homeobox C3a |
|
hoxa4a
|
ENSDARG00000103862 | homeobox A4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa3a | dr11_v1_chr19_+_19777437_19777437 | 0.29 | 5.2e-03 | Click! |
| hoxc3a | dr11_v1_chr23_+_36106790_36106790 | -0.14 | 1.9e-01 | Click! |
Activity profile of hoxa4a+hoxc3a motif
Sorted Z-values of hoxa4a+hoxc3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_20110086 | 7.31 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr19_-_5369486 | 4.42 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr22_-_10459880 | 4.20 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr5_-_71705191 | 3.79 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr15_-_12011390 | 3.58 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr4_+_73085993 | 3.54 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr21_-_32060993 | 3.42 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr11_-_1550709 | 3.24 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr2_+_11205795 | 3.23 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr15_-_46779934 | 3.22 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr5_+_56023186 | 3.20 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr6_-_54815886 | 3.14 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr10_-_22095505 | 3.03 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr7_+_31891110 | 2.99 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr22_+_19407531 | 2.74 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr17_+_16046314 | 2.71 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr25_+_29160102 | 2.69 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr12_-_464007 | 2.51 |
ENSDART00000106669
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr25_+_5035343 | 2.38 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr12_-_33357655 | 2.37 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr8_-_23780334 | 2.35 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr2_+_6253246 | 2.30 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr17_+_16046132 | 2.23 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr3_+_27798094 | 2.22 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr19_+_2631565 | 2.21 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr24_+_22731228 | 2.21 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr22_+_6254194 | 2.19 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr8_+_22516728 | 2.15 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr8_-_36287046 | 2.11 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr10_-_35257458 | 2.10 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr21_+_25777425 | 1.95 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr17_-_37395460 | 1.91 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr25_+_37293312 | 1.86 |
ENSDART00000086737
ENSDART00000161595 |
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr24_-_33291784 | 1.84 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr18_-_48550426 | 1.80 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr3_-_16784280 | 1.78 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr9_+_44994214 | 1.74 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr24_+_19415124 | 1.74 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr18_-_46258612 | 1.71 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr24_-_40744672 | 1.71 |
ENSDART00000160672
|
CU633479.1
|
|
| chr7_-_71389375 | 1.68 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr22_-_38621438 | 1.68 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr19_+_41464870 | 1.68 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr9_-_35633827 | 1.65 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr19_-_25149598 | 1.65 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr24_-_27452488 | 1.65 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr20_-_9462433 | 1.65 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr19_-_5103141 | 1.64 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_-_31107537 | 1.63 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr2_+_5371492 | 1.62 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr8_+_31717175 | 1.62 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr10_-_21362071 | 1.61 |
ENSDART00000125167
|
avd
|
avidin |
| chr6_+_28208973 | 1.60 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr10_-_21362320 | 1.60 |
ENSDART00000189789
|
avd
|
avidin |
| chr12_+_22580579 | 1.59 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr18_-_40708537 | 1.57 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr18_-_48547564 | 1.57 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr16_-_42056137 | 1.55 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr18_+_23408073 | 1.50 |
ENSDART00000136489
|
mctp2a
|
multiple C2 domains, transmembrane 2a |
| chr24_-_25166720 | 1.49 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr20_+_36812368 | 1.48 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr19_-_5103313 | 1.43 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr9_+_31795343 | 1.43 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr18_+_924949 | 1.43 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr17_+_16429826 | 1.40 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr22_-_30770751 | 1.38 |
ENSDART00000172115
|
AL831726.2
|
|
| chr23_+_28322986 | 1.38 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr22_-_10121880 | 1.38 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr12_+_23912074 | 1.37 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr16_-_20312146 | 1.32 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr14_+_46313396 | 1.31 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_+_34620418 | 1.30 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr19_+_46158078 | 1.30 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr20_-_37813863 | 1.27 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr3_+_18555899 | 1.27 |
ENSDART00000163036
ENSDART00000158791 |
cbx4
|
chromobox homolog 4 (Pc class homolog, Drosophila) |
| chr5_+_32206378 | 1.26 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr25_+_36292057 | 1.24 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr19_+_43297546 | 1.22 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr10_+_33393829 | 1.22 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr3_-_15734358 | 1.21 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr21_-_43015383 | 1.21 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr2_-_15324837 | 1.21 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr3_+_1182315 | 1.19 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr21_-_26490186 | 1.19 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr12_-_46176115 | 1.18 |
ENSDART00000152848
|
si:ch211-226h7.8
|
si:ch211-226h7.8 |
| chr12_-_48188928 | 1.17 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr20_+_29209615 | 1.17 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_-_23426339 | 1.16 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr3_-_34084387 | 1.16 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr20_-_29864390 | 1.15 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr1_+_53842344 | 1.14 |
ENSDART00000114506
|
crfb17
|
cytokine receptor family member B17 |
| chr5_+_63302660 | 1.13 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr16_+_23799622 | 1.13 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr22_+_19366866 | 1.13 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr22_+_737211 | 1.13 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr14_-_1355544 | 1.12 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr13_-_35808904 | 1.11 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr11_-_2131280 | 1.11 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr19_+_26923274 | 1.11 |
ENSDART00000148439
ENSDART00000148877 |
nelfe
|
negative elongation factor complex member E |
| chr10_+_15025006 | 1.09 |
ENSDART00000145192
ENSDART00000140084 |
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr17_+_8799451 | 1.09 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr9_+_38372216 | 1.09 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr23_-_36446307 | 1.08 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr24_-_9300160 | 1.08 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_+_20793982 | 1.08 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr9_+_30421489 | 1.08 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr10_+_15024772 | 1.07 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr15_-_43284021 | 1.06 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr10_+_44699734 | 1.06 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr6_+_7414215 | 1.05 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr5_-_4418555 | 1.05 |
ENSDART00000170158
|
apooa
|
apolipoprotein O, a |
| chr3_-_15734530 | 1.05 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr7_-_59165640 | 1.04 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr11_+_37250839 | 1.03 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr1_+_513986 | 1.03 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr2_+_36112273 | 0.99 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr4_+_77943184 | 0.99 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_-_16719244 | 0.98 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr8_-_11988065 | 0.98 |
ENSDART00000005140
|
med27
|
mediator complex subunit 27 |
| chr24_+_1023839 | 0.97 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr17_-_49978986 | 0.97 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr21_+_15592426 | 0.97 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr8_-_23776399 | 0.95 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr23_+_45966436 | 0.94 |
ENSDART00000172160
|
CABZ01069338.1
|
|
| chr13_+_9432501 | 0.93 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr16_-_51271962 | 0.93 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr23_+_2728095 | 0.92 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr19_+_4888491 | 0.91 |
ENSDART00000151540
|
cdk12
|
cyclin-dependent kinase 12 |
| chr22_-_20166660 | 0.91 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr16_+_54209504 | 0.90 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr20_+_29209767 | 0.89 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr1_-_55248496 | 0.89 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr13_+_4871886 | 0.88 |
ENSDART00000132301
|
micu1
|
mitochondrial calcium uptake 1 |
| chr11_-_20096018 | 0.86 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr10_+_44700103 | 0.86 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr6_+_21001264 | 0.85 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr22_-_21476836 | 0.85 |
ENSDART00000135977
|
s1pr3a
|
sphingosine-1-phosphate receptor 3a |
| chr5_+_66433287 | 0.85 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr3_-_27713180 | 0.84 |
ENSDART00000180151
|
CR394572.1
|
|
| chr13_+_33117528 | 0.84 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr10_-_2971407 | 0.83 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr6_+_16468776 | 0.82 |
ENSDART00000109151
ENSDART00000114667 |
zgc:161969
|
zgc:161969 |
| chr14_+_3038473 | 0.81 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr8_+_30664077 | 0.80 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr10_+_31951338 | 0.79 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr23_-_24394719 | 0.78 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr24_-_37640705 | 0.78 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr24_-_36680261 | 0.78 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr2_+_10007113 | 0.78 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr7_+_17816006 | 0.78 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr24_+_17260001 | 0.77 |
ENSDART00000066765
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr4_+_21129752 | 0.77 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr22_+_14051894 | 0.77 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr14_-_413273 | 0.76 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr6_-_55399214 | 0.75 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr21_-_37075542 | 0.75 |
ENSDART00000129439
|
BX649250.1
|
|
| chr14_-_25935167 | 0.74 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr5_+_54497730 | 0.73 |
ENSDART00000157722
|
tmem203
|
transmembrane protein 203 |
| chr21_-_14251306 | 0.73 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr11_-_11791718 | 0.72 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr7_-_28148310 | 0.71 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr8_-_38355268 | 0.70 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr1_-_51710225 | 0.70 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr19_-_5358443 | 0.70 |
ENSDART00000105036
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr15_+_12429206 | 0.70 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr5_-_3927692 | 0.69 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr3_-_12930217 | 0.69 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr3_+_1637358 | 0.69 |
ENSDART00000180266
|
CR394546.5
|
|
| chr24_+_16547035 | 0.69 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr8_+_17775247 | 0.69 |
ENSDART00000112356
|
si:ch211-150o23.3
|
si:ch211-150o23.3 |
| chr9_+_18829360 | 0.69 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr7_-_8052829 | 0.68 |
ENSDART00000172931
|
si:ch211-163c2.2
|
si:ch211-163c2.2 |
| chr24_+_36204028 | 0.68 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr7_+_22543963 | 0.68 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr9_-_710896 | 0.68 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr1_+_54952540 | 0.68 |
ENSDART00000182944
|
r3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr24_+_17260329 | 0.68 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr18_+_27571448 | 0.68 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr11_+_44356504 | 0.67 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr3_-_402714 | 0.67 |
ENSDART00000134062
ENSDART00000105659 |
mhc1zja
|
major histocompatibility complex class I ZJA |
| chr3_-_20040636 | 0.66 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr13_+_45431660 | 0.66 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr14_+_33329420 | 0.66 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr20_+_29209926 | 0.66 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr8_-_52229462 | 0.66 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr16_+_13818500 | 0.66 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr12_-_19119176 | 0.65 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr22_-_6229275 | 0.65 |
ENSDART00000146045
ENSDART00000179730 |
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr23_+_31107685 | 0.64 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr5_+_40485503 | 0.64 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr22_+_18156000 | 0.64 |
ENSDART00000143483
ENSDART00000136133 |
nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr7_+_36898850 | 0.63 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr6_+_6797520 | 0.63 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr21_+_28958471 | 0.62 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_+_24165371 | 0.62 |
ENSDART00000173472
|
si:ch211-216p19.5
|
si:ch211-216p19.5 |
| chr23_+_29885019 | 0.62 |
ENSDART00000167059
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr11_+_44356707 | 0.62 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr7_-_4125021 | 0.61 |
ENSDART00000167182
ENSDART00000173696 |
zgc:55733
|
zgc:55733 |
| chr9_-_27398369 | 0.60 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr12_-_44019561 | 0.59 |
ENSDART00000165900
|
si:dkey-31i7.1
|
si:dkey-31i7.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa4a+hoxc3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 1.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.6 | 1.9 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.6 | 3.0 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.5 | 1.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.4 | 2.9 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.4 | 2.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 1.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 1.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.3 | 1.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.3 | 0.8 | GO:0060907 | dendritic cell antigen processing and presentation(GO:0002468) macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.2 | 1.2 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.2 | 1.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 1.4 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.2 | 0.7 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 3.8 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.9 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 10.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.8 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.2 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 1.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 1.5 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.2 | 0.5 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.9 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 1.1 | GO:0072091 | forebrain anterior/posterior pattern specification(GO:0021797) regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.7 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 0.5 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.5 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 1.3 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.0 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.9 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.6 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.4 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 3.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 3.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 1.4 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.8 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.0 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.5 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.3 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 2.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 1.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.3 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.7 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.1 | 1.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 4.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 1.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.0 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 3.2 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.9 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.5 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.1 | 3.1 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 3.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 2.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 4.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 2.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.7 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.0 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.1 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 2.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 1.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 1.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.6 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.2 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 1.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.0 | GO:0051297 | centrosome cycle(GO:0007098) centrosome organization(GO:0051297) |
| 0.0 | 1.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.2 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.1 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.3 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 1.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 10.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 1.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 1.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.6 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.9 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 2.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 5.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 3.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 2.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 5.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.0 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 3.4 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.7 | 4.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 3.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.4 | 1.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 1.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 1.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.3 | 3.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 7.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 0.8 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 3.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.7 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.2 | 3.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 2.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 3.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 3.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.3 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.3 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.0 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.7 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.4 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.7 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 3.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 2.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.8 | GO:0019957 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 8.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 3.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.9 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.1 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |