Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
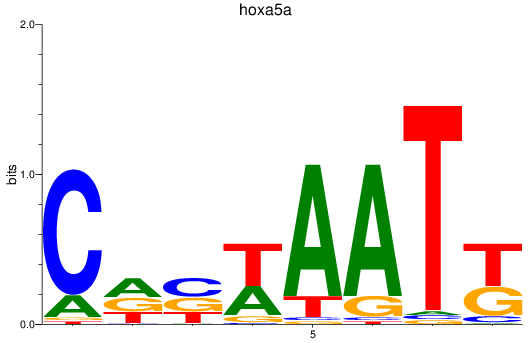
Results for hoxa5a
Z-value: 0.37
Transcription factors associated with hoxa5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa5a
|
ENSDARG00000102501 | homeobox A5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa5a | dr11_v1_chr19_+_19759577_19759577 | 0.12 | 2.5e-01 | Click! |
Activity profile of hoxa5a motif
Sorted Z-values of hoxa5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_34065573 | 2.11 |
ENSDART00000186946
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr13_+_27316632 | 1.70 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr14_-_30490763 | 1.63 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr3_-_62380146 | 1.36 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr7_-_69983462 | 1.34 |
ENSDART00000123380
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr5_-_71460556 | 1.34 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr13_+_27316934 | 1.31 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr15_-_28200049 | 1.27 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr24_+_11105786 | 1.18 |
ENSDART00000175182
|
prlh2
|
prolactin releasing hormone 2 |
| chr6_-_39475304 | 1.12 |
ENSDART00000022761
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr25_-_20523927 | 1.08 |
ENSDART00000114448
|
si:ch211-269c21.2
|
si:ch211-269c21.2 |
| chr3_-_30061985 | 1.06 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr12_+_28574863 | 1.02 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr21_+_11509461 | 0.98 |
ENSDART00000134620
|
entpd2b
|
ectonucleoside triphosphate diphosphohydrolase 2b |
| chr1_-_11850353 | 0.98 |
ENSDART00000033361
|
ttyh3b
|
tweety family member 3b |
| chr3_-_59981476 | 0.96 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr2_-_49978227 | 0.95 |
ENSDART00000142835
|
hecw1b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1b |
| chr20_+_52774730 | 0.94 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr8_+_47571211 | 0.94 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr25_-_23745765 | 0.90 |
ENSDART00000089278
|
si:ch211-236l14.4
|
si:ch211-236l14.4 |
| chr15_+_1372343 | 0.90 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr10_+_21576909 | 0.90 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr18_+_11506561 | 0.90 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr20_+_572037 | 0.87 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr10_+_39091353 | 0.82 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr22_-_20695237 | 0.81 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr8_-_18262149 | 0.80 |
ENSDART00000143000
|
rnf220b
|
ring finger protein 220b |
| chr9_+_52492639 | 0.78 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr15_-_16679517 | 0.76 |
ENSDART00000177384
|
caln1
|
calneuron 1 |
| chr8_-_36287046 | 0.76 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr13_-_32626247 | 0.74 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr8_-_50888806 | 0.71 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr2_+_37430917 | 0.71 |
ENSDART00000140558
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr14_-_2322484 | 0.69 |
ENSDART00000167806
|
si:ch73-379j16.2
|
si:ch73-379j16.2 |
| chr7_-_27685365 | 0.69 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr1_-_14332283 | 0.68 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr21_-_35325466 | 0.68 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr14_+_6159162 | 0.68 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr19_-_47570672 | 0.67 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr1_-_20273227 | 0.66 |
ENSDART00000146084
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr11_-_38083397 | 0.64 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr7_+_3357973 | 0.63 |
ENSDART00000172853
|
si:ch211-285c6.5
|
si:ch211-285c6.5 |
| chr19_+_19600297 | 0.63 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr7_-_55648336 | 0.63 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr6_-_39489190 | 0.63 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr19_+_712127 | 0.62 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr3_-_55404985 | 0.61 |
ENSDART00000154274
|
mafga
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
| chr7_-_41964877 | 0.60 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr23_-_31506854 | 0.59 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr11_+_18183220 | 0.56 |
ENSDART00000113468
|
LO018315.10
|
|
| chr9_-_34937025 | 0.56 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr10_+_5025696 | 0.56 |
ENSDART00000031165
|
enoph1
|
enolase-phosphatase 1 |
| chr16_-_23379464 | 0.56 |
ENSDART00000045891
|
trim46a
|
tripartite motif containing 46a |
| chr17_-_43558494 | 0.55 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr11_-_42980535 | 0.55 |
ENSDART00000181160
ENSDART00000192064 |
CABZ01069998.1
|
|
| chr24_+_11106402 | 0.55 |
ENSDART00000146697
|
prlh2
|
prolactin releasing hormone 2 |
| chr17_-_12966907 | 0.55 |
ENSDART00000022874
|
psma6a
|
proteasome subunit alpha 6a |
| chr21_+_22400951 | 0.53 |
ENSDART00000134592
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr4_-_10599062 | 0.52 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr4_-_20232974 | 0.52 |
ENSDART00000193353
|
stk38l
|
serine/threonine kinase 38 like |
| chr24_-_9300160 | 0.51 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr7_-_6604623 | 0.51 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr11_+_43751263 | 0.50 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr8_+_43053519 | 0.50 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr19_-_22568203 | 0.49 |
ENSDART00000182888
|
pleca
|
plectin a |
| chr23_+_32028574 | 0.48 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr15_-_47956388 | 0.48 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr18_+_1154189 | 0.47 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr2_-_33645411 | 0.46 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr11_-_16021424 | 0.46 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr12_+_3078221 | 0.45 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr3_+_17346502 | 0.45 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr8_+_35212233 | 0.44 |
ENSDART00000111021
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr13_+_29771463 | 0.44 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr21_-_4922981 | 0.43 |
ENSDART00000191819
ENSDART00000113556 ENSDART00000193269 ENSDART00000182961 |
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr23_+_40109353 | 0.43 |
ENSDART00000149249
|
ghrhrl
|
growth hormone releasing hormone receptor, like |
| chr3_+_24595922 | 0.42 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr19_-_41052408 | 0.42 |
ENSDART00000151120
|
sgce
|
sarcoglycan, epsilon |
| chr17_+_6828550 | 0.41 |
ENSDART00000153777
|
si:ch73-242m19.1
|
si:ch73-242m19.1 |
| chr21_+_35327589 | 0.40 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr13_-_17729474 | 0.40 |
ENSDART00000013011
|
vdac2
|
voltage-dependent anion channel 2 |
| chr2_+_13462305 | 0.40 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr7_-_33961551 | 0.39 |
ENSDART00000100104
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr2_+_56657804 | 0.39 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr15_-_15469079 | 0.39 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr8_-_30779101 | 0.38 |
ENSDART00000062229
|
p2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr21_-_2130241 | 0.38 |
ENSDART00000167648
ENSDART00000163781 |
si:rp71-1h20.5
|
si:rp71-1h20.5 |
| chr10_+_44700103 | 0.37 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr4_+_22480169 | 0.37 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr7_-_47850702 | 0.36 |
ENSDART00000109511
|
si:ch211-186j3.6
|
si:ch211-186j3.6 |
| chr5_+_59392183 | 0.36 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr4_-_2036620 | 0.36 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr2_+_6127593 | 0.36 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr19_+_21919856 | 0.35 |
ENSDART00000187306
ENSDART00000138544 |
galr1a
|
galanin receptor 1a |
| chr1_-_44000940 | 0.34 |
ENSDART00000134932
ENSDART00000138447 |
zgc:66472
|
zgc:66472 |
| chr17_-_43391499 | 0.34 |
ENSDART00000189280
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr3_-_32898626 | 0.32 |
ENSDART00000103201
|
kat7a
|
K(lysine) acetyltransferase 7a |
| chr22_+_19366866 | 0.32 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr7_-_40630698 | 0.32 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr9_+_44721808 | 0.32 |
ENSDART00000190578
|
nckap1
|
NCK-associated protein 1 |
| chr11_-_8782871 | 0.32 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr11_+_23810148 | 0.32 |
ENSDART00000127846
|
nfasca
|
neurofascin homolog (chicken) a |
| chr21_-_25668153 | 0.32 |
ENSDART00000192062
|
tmem179b
|
transmembrane protein 179B |
| chr5_-_20814576 | 0.31 |
ENSDART00000098682
ENSDART00000147639 |
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr23_-_16981899 | 0.31 |
ENSDART00000125472
ENSDART00000142297 ENSDART00000143180 |
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr13_-_12006007 | 0.31 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr21_+_45510448 | 0.30 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr10_+_44699734 | 0.29 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr17_-_8656155 | 0.29 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr5_+_39537195 | 0.28 |
ENSDART00000051256
|
fgf5
|
fibroblast growth factor 5 |
| chr21_-_2348838 | 0.28 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr5_+_31791001 | 0.28 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr24_-_31123365 | 0.27 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr11_+_25257022 | 0.27 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr1_-_10647307 | 0.26 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr11_-_3613558 | 0.26 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr5_+_31283576 | 0.26 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr13_-_37383625 | 0.25 |
ENSDART00000192888
ENSDART00000100322 ENSDART00000138934 |
kcnh5b
|
potassium voltage-gated channel, subfamily H (eag-related), member 5b |
| chr10_+_16069987 | 0.25 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr7_+_38809241 | 0.24 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr22_+_1700552 | 0.24 |
ENSDART00000166185
|
znf1154
|
zinc finger protein 1154 |
| chr24_-_21404367 | 0.23 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr10_-_1961576 | 0.23 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr9_-_1459985 | 0.23 |
ENSDART00000124777
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr18_+_41313999 | 0.23 |
ENSDART00000048985
|
veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr15_-_25613114 | 0.23 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr25_-_10622449 | 0.22 |
ENSDART00000155375
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr25_-_24233884 | 0.22 |
ENSDART00000146419
|
si:dkey-11e23.9
|
si:dkey-11e23.9 |
| chr15_+_897280 | 0.21 |
ENSDART00000155470
|
znf1012
|
zinc finger protein 1012 |
| chr7_+_6480212 | 0.21 |
ENSDART00000173320
|
si:cabz01036022.1
|
si:cabz01036022.1 |
| chr15_-_1844048 | 0.21 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_+_38913621 | 0.20 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr15_+_38887467 | 0.20 |
ENSDART00000185222
|
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr9_-_12269847 | 0.20 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr1_-_45614481 | 0.20 |
ENSDART00000148413
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_+_66433287 | 0.20 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr3_-_18410968 | 0.19 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr5_-_67783593 | 0.19 |
ENSDART00000010934
|
casr
|
calcium-sensing receptor |
| chr2_+_5300405 | 0.19 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr24_+_35981471 | 0.19 |
ENSDART00000157373
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr11_-_35575791 | 0.18 |
ENSDART00000031441
ENSDART00000188513 ENSDART00000183609 |
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr6_-_9236309 | 0.18 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr14_-_38826739 | 0.18 |
ENSDART00000187633
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr22_+_37631234 | 0.18 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr25_-_3627130 | 0.18 |
ENSDART00000171863
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr22_+_1462177 | 0.17 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr15_-_23692359 | 0.17 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr21_-_44772738 | 0.16 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr21_-_2299002 | 0.16 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr24_+_39277043 | 0.16 |
ENSDART00000165458
|
MPRIP
|
si:ch73-103b11.2 |
| chr14_-_16151055 | 0.16 |
ENSDART00000162431
|
ptcd3
|
pentatricopeptide repeat domain 3 |
| chr15_+_23799461 | 0.16 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr4_-_65037243 | 0.16 |
ENSDART00000170059
|
si:ch211-283l16.1
|
si:ch211-283l16.1 |
| chr21_-_2952789 | 0.16 |
ENSDART00000182898
|
znf971
|
zinc finger protein 971 |
| chr14_-_10762629 | 0.15 |
ENSDART00000061928
|
fgf16
|
fibroblast growth factor 16 |
| chr16_-_41059919 | 0.14 |
ENSDART00000188803
|
si:dkey-201i6.8
|
si:dkey-201i6.8 |
| chr12_-_31461412 | 0.13 |
ENSDART00000175929
ENSDART00000186342 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr1_-_11973341 | 0.13 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr13_-_12005429 | 0.13 |
ENSDART00000180302
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr14_-_33859149 | 0.13 |
ENSDART00000163877
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr8_-_11146687 | 0.13 |
ENSDART00000189626
|
asb13b
|
ankyrin repeat and SOCS box containing 13b |
| chr25_+_20694177 | 0.12 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr9_-_746317 | 0.12 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr7_+_34290051 | 0.12 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr18_+_30370339 | 0.11 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr23_-_6765653 | 0.11 |
ENSDART00000192310
|
FP102169.1
|
|
| chr7_-_24149670 | 0.11 |
ENSDART00000005884
|
mmp14a
|
matrix metallopeptidase 14a (membrane-inserted) |
| chr17_-_200316 | 0.10 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr22_+_17784414 | 0.09 |
ENSDART00000188189
ENSDART00000145260 |
tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr2_-_38204845 | 0.09 |
ENSDART00000142342
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr2_-_1468258 | 0.09 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr3_+_27606505 | 0.09 |
ENSDART00000150953
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr21_-_25618175 | 0.09 |
ENSDART00000133512
|
fosl1b
|
FOS-like antigen 1b |
| chr2_+_31597841 | 0.09 |
ENSDART00000162181
|
si:ch211-106h4.4
|
si:ch211-106h4.4 |
| chr16_-_25085327 | 0.08 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr2_+_32846602 | 0.08 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr22_+_2844865 | 0.08 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr13_-_8601184 | 0.08 |
ENSDART00000045086
|
prkceb
|
protein kinase C, epsilon b |
| chr16_-_2390931 | 0.08 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr7_+_23577723 | 0.08 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr16_-_48074540 | 0.07 |
ENSDART00000187258
ENSDART00000186694 |
CSMD3 (1 of many)
|
si:ch211-236p22.1 |
| chr4_+_57099307 | 0.07 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr7_-_25930594 | 0.06 |
ENSDART00000192834
|
BX005022.1
|
|
| chr2_+_32826235 | 0.06 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr2_-_43942595 | 0.06 |
ENSDART00000147925
|
si:ch211-195h23.4
|
si:ch211-195h23.4 |
| chr17_+_50248152 | 0.06 |
ENSDART00000153998
|
zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr6_-_2134581 | 0.06 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr2_+_37897079 | 0.06 |
ENSDART00000141784
|
tep1
|
telomerase-associated protein 1 |
| chr17_-_22324727 | 0.06 |
ENSDART00000160341
|
CU104709.1
|
|
| chr3_-_30388525 | 0.06 |
ENSDART00000183790
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr2_-_37803614 | 0.05 |
ENSDART00000154124
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr19_+_11432330 | 0.05 |
ENSDART00000188065
|
PQLC1
|
PQ loop repeat containing 1 |
| chr9_-_30220818 | 0.05 |
ENSDART00000140929
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr8_-_42594380 | 0.05 |
ENSDART00000140126
ENSDART00000135238 ENSDART00000192764 |
dok2
|
docking protein 2 |
| chr19_-_11248982 | 0.04 |
ENSDART00000091237
|
irgf3
|
immunity-related GTPase family, f3 |
| chr20_-_23041275 | 0.04 |
ENSDART00000188200
|
BX511028.1
|
|
| chr23_-_46034609 | 0.04 |
ENSDART00000158712
|
zgc:65873
|
zgc:65873 |
| chr8_-_4892913 | 0.04 |
ENSDART00000126808
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr16_+_23531583 | 0.03 |
ENSDART00000146708
|
adar
|
adenosine deaminase, RNA-specific |
| chr19_-_47832853 | 0.03 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.7 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.6 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.2 | 1.0 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.9 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 1.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.9 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.3 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 2.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.1 | 0.5 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.6 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.4 | GO:0021588 | cerebellum formation(GO:0021588) anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.5 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 3.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 | 0.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.9 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.3 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.4 | GO:0030800 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.2 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 1.1 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 1.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.2 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.5 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.6 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 0.6 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.3 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.7 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 3.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 1.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.8 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.6 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |