Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
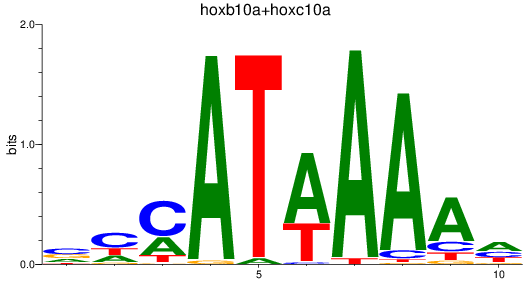
Results for hoxb10a+hoxc10a
Z-value: 0.76
Transcription factors associated with hoxb10a+hoxc10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb10a
|
ENSDARG00000011579 | homeobox B10a |
|
hoxc10a
|
ENSDARG00000070348 | homeobox C10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb10a | dr11_v1_chr3_+_23669267_23669267 | -0.21 | 4.3e-02 | Click! |
| hoxc10a | dr11_v1_chr23_+_36083529_36083617 | -0.20 | 5.1e-02 | Click! |
Activity profile of hoxb10a+hoxc10a motif
Sorted Z-values of hoxb10a+hoxc10a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_20373058 | 8.35 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr1_-_38195012 | 7.37 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr6_-_30485009 | 7.02 |
ENSDART00000025698
|
zgc:153311
|
zgc:153311 |
| chr20_+_26538137 | 6.73 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr6_+_36877968 | 6.32 |
ENSDART00000155187
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr14_+_30279391 | 6.26 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr8_+_3379815 | 5.87 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr24_-_27473771 | 5.49 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr10_+_26612321 | 5.13 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr23_+_39606108 | 4.91 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr24_-_27452488 | 4.77 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr5_+_15202495 | 4.76 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr23_+_10146542 | 4.73 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr4_+_7391110 | 4.66 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr13_-_12660318 | 4.58 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr21_-_25756119 | 4.50 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr17_+_27456804 | 4.27 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr7_+_48761875 | 4.12 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr25_+_4750972 | 3.98 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr4_+_7391400 | 3.90 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr19_+_2590182 | 3.87 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr16_-_45917322 | 3.69 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr19_+_19762183 | 3.66 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr11_-_6265574 | 3.49 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr12_-_22400999 | 3.48 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr7_+_22688781 | 3.45 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr5_-_24174956 | 3.39 |
ENSDART00000137631
|
si:ch211-137i24.10
|
si:ch211-137i24.10 |
| chr17_+_45395846 | 3.38 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr23_+_17417539 | 3.38 |
ENSDART00000182605
|
BX649300.2
|
|
| chr19_-_15855427 | 3.37 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr21_-_2415808 | 3.36 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr20_-_32045057 | 3.35 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr13_-_50108337 | 3.35 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr16_-_21038015 | 3.28 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr5_-_26181863 | 3.28 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr13_-_42749916 | 3.04 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr6_+_13045885 | 2.99 |
ENSDART00000104757
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr19_-_47571456 | 2.96 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr16_-_7443388 | 2.85 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr24_-_27461295 | 2.81 |
ENSDART00000110748
|
ccl34b.9
|
chemokine (C-C motif) ligand 34b, duplicate 9 |
| chr7_-_16562200 | 2.79 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr15_-_4580763 | 2.75 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr13_-_35984530 | 2.74 |
ENSDART00000143488
|
si:ch211-67f13.8
|
si:ch211-67f13.8 |
| chr23_+_7379728 | 2.73 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr1_-_37377509 | 2.65 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr23_+_24085531 | 2.56 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr12_+_46543572 | 2.55 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr1_+_1915967 | 2.54 |
ENSDART00000131463
|
si:ch211-132g1.1
|
si:ch211-132g1.1 |
| chr5_-_57723929 | 2.52 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr24_+_12913329 | 2.44 |
ENSDART00000141829
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr16_+_50969248 | 2.33 |
ENSDART00000172068
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr16_+_25535993 | 2.29 |
ENSDART00000077436
|
mylipb
|
myosin regulatory light chain interacting protein b |
| chr20_+_6543674 | 2.07 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr14_-_31488100 | 2.00 |
ENSDART00000186246
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr6_+_27624023 | 1.98 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr17_+_33453689 | 1.98 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr19_+_17385561 | 1.95 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr7_+_34296789 | 1.90 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr18_-_17513426 | 1.87 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr3_+_35812040 | 1.86 |
ENSDART00000075903
ENSDART00000147712 |
crlf3
|
cytokine receptor-like factor 3 |
| chr10_+_36650222 | 1.85 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr7_-_60096318 | 1.80 |
ENSDART00000189125
|
BX511067.1
|
|
| chr11_-_18253111 | 1.80 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr20_+_52546186 | 1.80 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr7_+_60111581 | 1.78 |
ENSDART00000087093
|
hspa12b
|
heat shock protein 12B |
| chr25_+_29474982 | 1.77 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr8_+_2487250 | 1.76 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr13_+_24584401 | 1.71 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr6_+_41191482 | 1.69 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr3_+_46764278 | 1.69 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr20_-_29499363 | 1.68 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr20_-_33961697 | 1.67 |
ENSDART00000061765
|
selp
|
selectin P |
| chr16_+_25664387 | 1.59 |
ENSDART00000184394
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr19_+_19761966 | 1.57 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr6_-_28222592 | 1.54 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr8_-_19487327 | 1.54 |
ENSDART00000111710
|
edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr7_+_58179513 | 1.54 |
ENSDART00000123117
|
ggh
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr23_-_43718067 | 1.52 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr4_-_14191717 | 1.52 |
ENSDART00000147928
|
pus7l
|
pseudouridylate synthase 7-like |
| chr20_-_33966148 | 1.51 |
ENSDART00000148111
|
selp
|
selectin P |
| chr10_+_2587234 | 1.48 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr4_+_51552434 | 1.47 |
ENSDART00000165016
|
si:dkey-165e24.1
|
si:dkey-165e24.1 |
| chr16_+_25664078 | 1.47 |
ENSDART00000154024
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr7_+_24165371 | 1.45 |
ENSDART00000173472
|
si:ch211-216p19.5
|
si:ch211-216p19.5 |
| chr20_-_25533739 | 1.44 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr5_-_36597612 | 1.43 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr6_+_30703828 | 1.42 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr2_-_55298075 | 1.40 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr4_-_14191434 | 1.39 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr10_-_35002731 | 1.37 |
ENSDART00000131279
|
alg5
|
asparagine-linked glycosylation 5 (dolichyl-phosphate beta-glucosyltransferase) |
| chr16_-_41646164 | 1.36 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr18_-_15329454 | 1.32 |
ENSDART00000191903
ENSDART00000019818 |
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr23_+_25354856 | 1.31 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr2_-_6039757 | 1.31 |
ENSDART00000013079
|
scp2a
|
sterol carrier protein 2a |
| chr11_+_14284866 | 1.26 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr21_-_32781612 | 1.20 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr1_-_9249943 | 1.19 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr10_+_5234327 | 1.18 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr5_-_54712159 | 1.16 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr17_+_17764979 | 1.15 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr18_+_41232719 | 1.14 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr13_-_26799244 | 1.11 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr2_-_28420415 | 1.10 |
ENSDART00000183857
|
CABZ01056051.1
|
|
| chr19_-_47571797 | 1.06 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_+_28187480 | 1.01 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr4_-_14192254 | 0.99 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr2_+_52847049 | 0.97 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr14_+_23717165 | 0.96 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr14_+_21159693 | 0.95 |
ENSDART00000192678
|
zgc:136929
|
zgc:136929 |
| chr1_-_35924495 | 0.94 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr2_-_35566938 | 0.91 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr23_+_27068225 | 0.91 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr1_+_51407520 | 0.89 |
ENSDART00000074294
|
actr2a
|
ARP2 actin related protein 2a homolog |
| chr17_+_47090497 | 0.87 |
ENSDART00000169038
ENSDART00000159292 |
zgc:103755
|
zgc:103755 |
| chr25_-_31898552 | 0.87 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr11_+_36683859 | 0.85 |
ENSDART00000170102
|
si:ch211-11c3.12
|
si:ch211-11c3.12 |
| chr4_-_56898328 | 0.85 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr10_+_44692272 | 0.83 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr12_-_13155653 | 0.82 |
ENSDART00000152467
|
cxl34c
|
CX chemokine ligand 34c |
| chr22_-_4644484 | 0.81 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr25_-_13614863 | 0.77 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr23_+_43718115 | 0.76 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr25_+_20089986 | 0.76 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr9_+_38588081 | 0.75 |
ENSDART00000031127
ENSDART00000131784 |
snx4
|
sorting nexin 4 |
| chr2_-_38287987 | 0.74 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr21_-_14826066 | 0.72 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr4_+_2267641 | 0.70 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr5_-_23715861 | 0.70 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr15_-_1843831 | 0.70 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr12_+_19191787 | 0.69 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr2_+_35595454 | 0.67 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr8_+_2487883 | 0.67 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr6_+_13083146 | 0.65 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr6_+_35362225 | 0.65 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr20_+_16173618 | 0.64 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr16_+_13883872 | 0.63 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr13_-_50546634 | 0.62 |
ENSDART00000192127
|
CU570781.2
|
|
| chr5_-_42883761 | 0.61 |
ENSDART00000167374
|
BX323596.2
|
|
| chr13_+_45431660 | 0.60 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr19_+_7759354 | 0.57 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr7_-_41338923 | 0.55 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr2_+_24374305 | 0.53 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr3_-_31069776 | 0.52 |
ENSDART00000167462
|
elob
|
elongin B |
| chr23_-_9807546 | 0.51 |
ENSDART00000136740
ENSDART00000004474 |
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr12_-_13337033 | 0.51 |
ENSDART00000105903
ENSDART00000139786 |
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr20_-_51186524 | 0.51 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr13_-_37519774 | 0.51 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr1_+_2301961 | 0.51 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr17_-_47090440 | 0.48 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr3_+_32933663 | 0.47 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr8_+_18588551 | 0.47 |
ENSDART00000177476
ENSDART00000063539 |
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr7_+_27317174 | 0.45 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr24_+_1042594 | 0.44 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr13_+_9612395 | 0.44 |
ENSDART00000136689
ENSDART00000138362 |
slf2
|
SMC5-SMC6 complex localization factor 2 |
| chr20_+_9763364 | 0.43 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr15_-_28860282 | 0.41 |
ENSDART00000156738
|
gpr4
|
G protein-coupled receptor 4 |
| chr21_-_26520629 | 0.41 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr14_-_33613794 | 0.40 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr5_+_30588856 | 0.40 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr6_-_37749711 | 0.38 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr13_+_22295905 | 0.37 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr8_+_30709685 | 0.35 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr19_-_10881486 | 0.35 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr24_-_12689571 | 0.35 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr23_-_24047054 | 0.35 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr17_+_47116500 | 0.32 |
ENSDART00000186627
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr12_+_31638045 | 0.32 |
ENSDART00000184216
ENSDART00000183645 ENSDART00000153129 |
dnmbp
|
dynamin binding protein |
| chr17_-_2690083 | 0.31 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr19_-_22843480 | 0.31 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr12_-_26430507 | 0.30 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr21_+_20396858 | 0.30 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr21_-_27251112 | 0.30 |
ENSDART00000137667
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr12_+_34051848 | 0.29 |
ENSDART00000153276
|
cyth1b
|
cytohesin 1b |
| chr11_-_24016761 | 0.28 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr3_-_32873641 | 0.28 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr7_-_69025306 | 0.25 |
ENSDART00000180796
|
CABZ01057488.2
|
|
| chr6_-_39653972 | 0.24 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr2_-_45663945 | 0.24 |
ENSDART00000075080
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr4_+_44954486 | 0.23 |
ENSDART00000150875
|
si:dkeyp-100h4.8
|
si:dkeyp-100h4.8 |
| chr10_-_2713228 | 0.23 |
ENSDART00000123754
ENSDART00000126236 |
mier3a
|
mesoderm induction early response 1, family member 3 a |
| chr5_-_50371205 | 0.22 |
ENSDART00000157800
|
si:ch73-280o22.2
|
si:ch73-280o22.2 |
| chr12_-_19091214 | 0.22 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr6_-_12588044 | 0.21 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr21_+_4313039 | 0.21 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr22_+_1779401 | 0.20 |
ENSDART00000170126
|
znf1154
|
zinc finger protein 1154 |
| chr18_+_7363242 | 0.16 |
ENSDART00000133375
|
si:dkey-30c15.17
|
si:dkey-30c15.17 |
| chr9_-_1939232 | 0.16 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr1_-_39983730 | 0.16 |
ENSDART00000160066
|
ing2
|
inhibitor of growth family, member 2 |
| chr14_-_30967284 | 0.15 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr22_-_8594846 | 0.15 |
ENSDART00000081690
ENSDART00000192241 |
si:ch73-27e22.1
|
si:ch73-27e22.1 |
| chr10_-_10969444 | 0.14 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr16_+_39271123 | 0.13 |
ENSDART00000043823
ENSDART00000141801 |
osbpl10b
|
oxysterol binding protein-like 10b |
| chr5_-_57289872 | 0.12 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr15_-_5212930 | 0.11 |
ENSDART00000136845
|
or128-6
|
odorant receptor, family E, subfamily 128, member 6 |
| chr14_+_23593989 | 0.11 |
ENSDART00000163682
|
med12
|
mediator complex subunit 12 |
| chr7_-_29341233 | 0.10 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr8_+_8860902 | 0.10 |
ENSDART00000144986
|
otud5a
|
OTU deubiquitinase 5a |
| chr1_+_40801448 | 0.10 |
ENSDART00000187394
|
cpz
|
carboxypeptidase Z |
| chr12_+_17603528 | 0.10 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr5_+_24245682 | 0.09 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb10a+hoxc10a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 1.2 | 7.4 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 1.2 | 3.5 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.0 | 3.0 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.9 | 2.6 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.7 | 5.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.7 | 2.8 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.7 | 4.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.7 | 2.7 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.7 | 4.6 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.5 | 3.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.5 | 5.7 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.4 | 2.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 2.8 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 | 1.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 3.4 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.4 | 1.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.4 | 1.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.3 | 3.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.3 | 1.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 1.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 0.8 | GO:2000136 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.3 | 1.3 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.3 | 3.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 1.9 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 1.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.2 | 5.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.2 | 2.0 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.2 | 6.3 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.2 | 3.3 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 1.1 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.2 | 9.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 7.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 3.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.2 | 1.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.9 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.5 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.4 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 1.9 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.7 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 0.9 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 1.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 3.7 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.9 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 3.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 6.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 0.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 1.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.5 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 4.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.4 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 2.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.4 | GO:1902622 | Rac protein signal transduction(GO:0016601) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.7 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 4.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 1.4 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 1.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 2.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 10.6 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.9 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.2 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.4 | 3.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.4 | 1.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.4 | 1.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.3 | 6.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 1.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.7 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 9.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 3.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.8 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 5.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 6.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 31.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.9 | 4.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.8 | 5.7 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 1.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 1.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.5 | 1.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 7.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.3 | 1.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.2 | 0.7 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.2 | 3.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 1.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 3.0 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 9.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 3.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.5 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.2 | 2.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.5 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.9 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 1.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 2.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.1 | 4.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 5.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 3.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 2.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 4.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 4.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 3.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 3.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 4.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 5.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 5.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 2.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 5.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 3.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 1.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 2.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |