Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
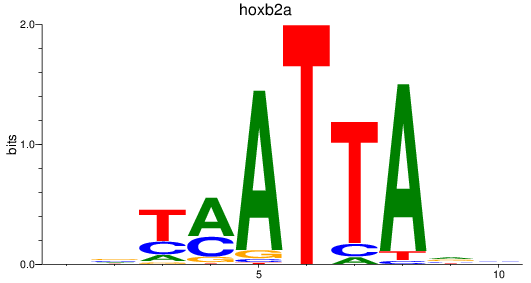
Results for hoxb2a
Z-value: 0.74
Transcription factors associated with hoxb2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb2a
|
ENSDARG00000000175 | homeobox B2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb2a | dr11_v1_chr3_+_23752150_23752150 | 0.37 | 2.4e-04 | Click! |
Activity profile of hoxb2a motif
Sorted Z-values of hoxb2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_9462433 | 7.66 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr9_+_38372216 | 7.05 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr8_+_22930627 | 6.16 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr16_+_5774977 | 5.24 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr23_-_26522760 | 5.05 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr18_+_17428258 | 4.84 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr11_+_30057762 | 4.83 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr9_-_31278048 | 4.43 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr5_+_64732036 | 4.25 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr3_+_1637358 | 4.23 |
ENSDART00000180266
|
CR394546.5
|
|
| chr4_-_16833518 | 4.19 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr6_-_54826061 | 4.12 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr1_+_16397063 | 4.01 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr17_+_23298928 | 3.96 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr19_-_31402429 | 3.77 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr19_+_14921000 | 3.63 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr21_-_39177564 | 3.52 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr21_+_28958471 | 3.48 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_45365098 | 3.40 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr22_-_20166660 | 3.31 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr23_-_14990865 | 3.27 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr13_+_15838151 | 3.26 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr22_-_881080 | 3.25 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr1_-_10647484 | 3.25 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr14_+_24935131 | 3.22 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr22_+_1796057 | 3.22 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr1_-_10647307 | 3.20 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr22_+_17205608 | 3.17 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr5_+_64732270 | 3.03 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr17_+_31185276 | 3.03 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr1_-_50859053 | 3.02 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr9_+_38983895 | 3.01 |
ENSDART00000144893
|
map2
|
microtubule-associated protein 2 |
| chr24_+_16547035 | 3.01 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr1_+_513986 | 2.99 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr16_-_29458806 | 2.97 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr20_-_46362606 | 2.91 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr3_+_45364849 | 2.90 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr7_+_30787903 | 2.87 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr25_+_16116740 | 2.87 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr25_-_27722614 | 2.71 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr9_-_32753535 | 2.70 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr5_-_41494831 | 2.64 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr1_+_34696503 | 2.60 |
ENSDART00000186106
|
CR339054.2
|
|
| chr20_+_27020201 | 2.57 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr15_-_755023 | 2.56 |
ENSDART00000155594
|
znf1011
|
zinc finger protein 1011 |
| chr20_+_19512727 | 2.53 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr3_+_30922947 | 2.52 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr17_-_28198099 | 2.51 |
ENSDART00000156143
|
htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr15_-_9272328 | 2.51 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr24_+_17007407 | 2.51 |
ENSDART00000110652
|
zfx
|
zinc finger protein, X-linked |
| chr19_-_30524952 | 2.50 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr5_+_43965078 | 2.47 |
ENSDART00000113502
ENSDART00000187143 |
TMEM8B
|
si:dkey-84j12.1 |
| chr14_-_4273396 | 2.47 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr21_+_26726936 | 2.45 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr23_+_11285662 | 2.45 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr15_-_44601331 | 2.44 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr16_+_25011994 | 2.43 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr2_+_33326522 | 2.42 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr1_+_40562540 | 2.42 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr3_-_25814097 | 2.40 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr16_+_13818743 | 2.39 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr23_+_40460333 | 2.38 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr3_-_34337969 | 2.37 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr17_+_24722646 | 2.33 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr25_+_30298377 | 2.32 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr12_+_23912074 | 2.30 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr16_+_7242610 | 2.29 |
ENSDART00000081477
|
sri
|
sorcin |
| chr2_-_33687214 | 2.27 |
ENSDART00000147439
|
atp6v0b
|
ATPase H+ transporting V0 subunit b |
| chr2_+_50608099 | 2.24 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr11_+_14937904 | 2.24 |
ENSDART00000185103
|
CR550302.3
|
|
| chr3_+_33341640 | 2.23 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr20_-_38787341 | 2.22 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr1_+_16396870 | 2.22 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr2_+_27010439 | 2.19 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr18_+_18000887 | 2.19 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr22_-_11137268 | 2.18 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr2_+_20332044 | 2.18 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr13_-_40120252 | 2.17 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr19_-_5103141 | 2.16 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr14_+_23717165 | 2.15 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr10_-_34772211 | 2.13 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr15_+_6109861 | 2.12 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr14_+_30795559 | 2.10 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr4_+_4849789 | 2.09 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr1_-_45039726 | 2.06 |
ENSDART00000186188
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr7_+_14291323 | 2.06 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr20_-_35040041 | 2.03 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr7_-_25895189 | 2.01 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr5_+_63302660 | 1.98 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr24_-_1341543 | 1.97 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr17_-_25649079 | 1.97 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr3_-_32596394 | 1.94 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr3_+_34919810 | 1.93 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr3_+_22442445 | 1.92 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr20_-_27225876 | 1.91 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr25_+_28825657 | 1.91 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr5_-_26765188 | 1.91 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr25_+_35553542 | 1.91 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr8_+_28452738 | 1.90 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr17_-_31695217 | 1.89 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr12_+_47162456 | 1.89 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr8_-_46894362 | 1.88 |
ENSDART00000111124
|
acot7
|
acyl-CoA thioesterase 7 |
| chr16_+_21330634 | 1.87 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr20_-_45661049 | 1.86 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr7_+_7511914 | 1.85 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr25_-_27722309 | 1.84 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr19_-_5805923 | 1.84 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr1_+_54865552 | 1.83 |
ENSDART00000145381
|
si:ch211-196h16.5
|
si:ch211-196h16.5 |
| chr22_-_19552796 | 1.83 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr14_-_7137808 | 1.83 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr24_+_12989727 | 1.83 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr21_+_34088377 | 1.81 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr16_+_47207691 | 1.80 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr1_-_57280585 | 1.79 |
ENSDART00000152220
|
si:dkey-27j5.5
|
si:dkey-27j5.5 |
| chr18_-_2433011 | 1.79 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr12_+_24342303 | 1.75 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr1_+_11977426 | 1.74 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr18_+_18000695 | 1.74 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr23_-_24493768 | 1.74 |
ENSDART00000135559
ENSDART00000046933 |
sult1st5
|
sulfotransferase family 1, cytosolic sulfotransferase 5 |
| chr13_+_30903816 | 1.73 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr10_+_1849874 | 1.72 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr8_-_19216657 | 1.72 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr19_+_2631565 | 1.71 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr12_-_30500803 | 1.69 |
ENSDART00000153108
|
nhlrc2
|
NHL repeat containing 2 |
| chr23_-_36446307 | 1.69 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr21_+_34088110 | 1.66 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr1_+_40023640 | 1.65 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr17_+_24687338 | 1.65 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr21_-_44565236 | 1.64 |
ENSDART00000159323
|
fundc2
|
fun14 domain containing 2 |
| chr9_+_17983463 | 1.62 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr11_+_38280454 | 1.62 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr17_-_16422654 | 1.61 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr18_-_18875308 | 1.60 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr13_-_45475289 | 1.58 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr10_-_4375190 | 1.57 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr1_+_55293424 | 1.57 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr17_-_31639845 | 1.56 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr24_-_6078222 | 1.55 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr15_-_1844048 | 1.55 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr3_+_46459540 | 1.55 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
| chr14_+_12178915 | 1.52 |
ENSDART00000054626
|
hdac3
|
histone deacetylase 3 |
| chr17_-_22324727 | 1.52 |
ENSDART00000160341
|
CU104709.1
|
|
| chr6_-_14292307 | 1.52 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr7_+_71547747 | 1.51 |
ENSDART00000180869
|
adcyap1a
|
adenylate cyclase activating polypeptide 1a |
| chr19_-_7495006 | 1.51 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr12_-_3077395 | 1.50 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_+_54209504 | 1.48 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr7_-_72261721 | 1.47 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr3_+_32832538 | 1.46 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr3_-_32169754 | 1.46 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr7_-_71389375 | 1.45 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr7_-_40145097 | 1.45 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr14_-_8271686 | 1.44 |
ENSDART00000165120
|
purab
|
purine-rich element binding protein Ab |
| chr22_-_15593824 | 1.44 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr5_-_30615901 | 1.44 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr1_+_33647682 | 1.44 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr22_+_39096911 | 1.43 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr22_+_30496360 | 1.43 |
ENSDART00000189145
|
BX649490.4
|
|
| chr20_-_32270866 | 1.41 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr25_-_1235457 | 1.41 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr19_+_366034 | 1.40 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr18_-_29896367 | 1.40 |
ENSDART00000191303
|
cmc2
|
C-x(9)-C motif containing 2 |
| chr4_+_69559692 | 1.39 |
ENSDART00000164383
|
znf993
|
zinc finger protein 993 |
| chr9_+_41156818 | 1.38 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr22_+_2937485 | 1.38 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr7_-_7676186 | 1.37 |
ENSDART00000091105
|
mfap3l
|
microfibril associated protein 3 like |
| chr17_+_12658411 | 1.36 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr24_+_39518774 | 1.34 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr8_+_23355484 | 1.34 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr6_-_41138854 | 1.33 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr22_-_881725 | 1.30 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr21_-_3770636 | 1.30 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr5_-_55933420 | 1.30 |
ENSDART00000050966
|
slc25a46
|
solute carrier family 25, member 46 |
| chr11_-_18604542 | 1.30 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr8_-_37249991 | 1.29 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr5_-_50640171 | 1.28 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr24_-_39518599 | 1.27 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr24_+_23716918 | 1.24 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr11_+_28476298 | 1.23 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr2_+_56213694 | 1.23 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr16_+_9400661 | 1.23 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr1_+_52929185 | 1.21 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr12_+_16087077 | 1.21 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr6_-_40922971 | 1.21 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr16_+_1353894 | 1.21 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr14_+_8940326 | 1.21 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr17_-_21162821 | 1.20 |
ENSDART00000157283
|
abhd12
|
abhydrolase domain containing 12 |
| chr9_+_38163876 | 1.20 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr10_-_21955231 | 1.19 |
ENSDART00000183695
|
FO744833.2
|
|
| chr5_-_16734060 | 1.19 |
ENSDART00000035859
ENSDART00000190457 ENSDART00000179244 |
atad1a
|
ATPase family, AAA domain containing 1a |
| chr22_+_30543437 | 1.17 |
ENSDART00000137983
|
si:dkey-103k4.1
|
si:dkey-103k4.1 |
| chr3_-_60827402 | 1.17 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr4_-_193762 | 1.17 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr21_+_39365920 | 1.16 |
ENSDART00000140940
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr15_+_7086327 | 1.15 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr6_-_43283122 | 1.15 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr23_-_36418708 | 1.14 |
ENSDART00000132273
|
znf740b
|
zinc finger protein 740b |
| chr10_-_41400049 | 1.14 |
ENSDART00000009838
|
gpat4
|
glycerol-3-phosphate acyltransferase 4 |
| chr24_+_32592748 | 1.14 |
ENSDART00000188256
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr20_-_38787047 | 1.14 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.9 | 3.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.9 | 2.6 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.8 | 7.3 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.8 | 2.4 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.8 | 2.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.7 | 2.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.7 | 2.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.6 | 4.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.6 | 2.5 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.6 | 3.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.5 | 2.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.5 | 2.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.5 | 1.9 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.4 | 1.7 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.4 | 3.3 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.4 | 2.0 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.4 | 1.9 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.4 | 4.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 6.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 2.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 1.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.3 | 1.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 0.8 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 2.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 1.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 1.7 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.2 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 1.2 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.2 | 1.7 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.2 | 3.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 0.7 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 3.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 0.6 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 0.8 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 1.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 1.0 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 1.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 0.8 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 1.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 1.8 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.5 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 1.9 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 1.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.5 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 2.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 2.4 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.5 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.9 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.7 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 1.6 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.1 | 5.0 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 2.2 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.1 | 2.5 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 1.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 2.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 2.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 3.0 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.1 | 3.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 8.9 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 0.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 3.3 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.1 | 1.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 3.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 3.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 2.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.9 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 3.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.8 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.4 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 1.0 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.4 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.1 | 1.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 4.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.9 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.4 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 1.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.6 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 1.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.4 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.9 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 1.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 2.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.5 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 1.0 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 1.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.3 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 2.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 5.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 3.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 1.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 2.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.4 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.7 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.9 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 2.5 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.2 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 1.2 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 2.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 2.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.0 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 1.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.4 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.6 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) |
| 0.0 | 1.2 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 2.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.7 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.3 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 6.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.6 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 2.7 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.9 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 6.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.5 | 1.9 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 2.2 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.4 | 1.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 3.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 4.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.3 | 2.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 2.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 2.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 2.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 7.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.9 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.4 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 7.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 2.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 4.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 3.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 4.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 2.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 1.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 3.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 7.3 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 2.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 16.5 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.8 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 13.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.8 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 1.2 | 3.6 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.9 | 4.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.6 | 3.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 2.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 3.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.5 | 1.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.4 | 2.2 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.4 | 2.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 2.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 2.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.4 | 1.5 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.4 | 1.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 3.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 0.9 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.3 | 1.9 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 5.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.3 | 1.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 0.8 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 1.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 1.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 1.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.0 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.2 | 0.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.6 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 6.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 2.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.5 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.2 | 2.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 2.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 1.7 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 6.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.4 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 3.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 2.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 2.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 2.5 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 2.0 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 2.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.8 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 2.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 2.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 3.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 1.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.9 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.4 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 8.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 2.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 2.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 2.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 3.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 1.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 1.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |