Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
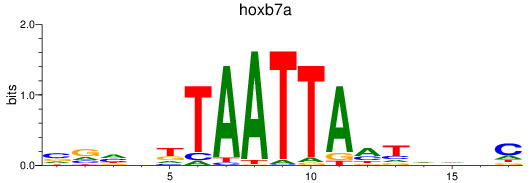
Results for hoxb7a
Z-value: 0.37
Transcription factors associated with hoxb7a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb7a
|
ENSDARG00000056030 | homeobox B7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb7a | dr11_v1_chr3_+_23691847_23691847 | 0.40 | 5.3e-05 | Click! |
Activity profile of hoxb7a motif
Sorted Z-values of hoxb7a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_7048245 | 3.59 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr25_+_14087045 | 2.82 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr13_+_10945337 | 2.64 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr12_-_35830625 | 2.24 |
ENSDART00000180028
|
CU459056.1
|
|
| chr2_-_1486023 | 2.21 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr13_-_10945288 | 2.20 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr7_+_69019851 | 2.07 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr10_-_21362320 | 1.94 |
ENSDART00000189789
|
avd
|
avidin |
| chr18_+_45781516 | 1.93 |
ENSDART00000168840
|
rpl35a
|
ribosomal protein L35a |
| chr24_+_16985181 | 1.91 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr10_-_21362071 | 1.86 |
ENSDART00000125167
|
avd
|
avidin |
| chr18_+_45781115 | 1.82 |
ENSDART00000151699
ENSDART00000179887 |
rpl35a
|
ribosomal protein L35a |
| chr12_+_20641471 | 1.79 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr12_+_20641102 | 1.73 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr17_+_16090436 | 1.55 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr5_+_34623107 | 1.54 |
ENSDART00000184126
|
enc1
|
ectodermal-neural cortex 1 |
| chr23_+_4709607 | 1.50 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr10_-_13343831 | 1.49 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr13_-_12602920 | 1.44 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr10_+_11261576 | 1.43 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr4_-_9891874 | 1.37 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr7_+_21272833 | 1.33 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr6_-_43677125 | 1.30 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr17_+_20589553 | 1.29 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr17_+_30369396 | 1.25 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr1_+_19303241 | 1.21 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr5_+_71802014 | 1.21 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr4_+_77971104 | 1.21 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr18_+_14619544 | 1.18 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr5_-_30620625 | 1.17 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr25_-_35599887 | 1.14 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr12_+_47698356 | 1.11 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr22_-_22301672 | 1.04 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr22_-_27296889 | 0.95 |
ENSDART00000155724
|
si:dkey-208m12.3
|
si:dkey-208m12.3 |
| chr12_+_48803098 | 0.92 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr9_-_27398369 | 0.91 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr12_+_16168342 | 0.90 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr4_-_779796 | 0.88 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr25_+_13620555 | 0.88 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr20_-_9095105 | 0.84 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr6_+_24398907 | 0.84 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr24_+_13316737 | 0.83 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr8_-_12867128 | 0.83 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr18_-_15551360 | 0.81 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr9_+_1139378 | 0.80 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr11_+_705727 | 0.79 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr10_-_31015535 | 0.78 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr13_-_2440622 | 0.77 |
ENSDART00000102767
ENSDART00000172616 |
fbxo9
|
F-box protein 9 |
| chr3_-_50443607 | 0.74 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr20_-_46128590 | 0.73 |
ENSDART00000123744
|
taar1b
|
trace amine associated receptor 1b |
| chr2_+_105748 | 0.71 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr17_+_51682429 | 0.70 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr15_+_28318005 | 0.70 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr23_+_28809002 | 0.70 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr24_+_7800486 | 0.69 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr6_+_7414215 | 0.65 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr4_-_13548806 | 0.64 |
ENSDART00000067155
|
il22
|
interleukin 22 |
| chr13_-_5252559 | 0.61 |
ENSDART00000181652
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr12_-_4243268 | 0.58 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr11_-_45141309 | 0.58 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr8_+_25034544 | 0.57 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr22_-_22301929 | 0.49 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr4_+_16715267 | 0.48 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr17_+_31914877 | 0.47 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr15_-_40246396 | 0.47 |
ENSDART00000063777
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr23_+_39461989 | 0.46 |
ENSDART00000184761
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr6_+_9898683 | 0.41 |
ENSDART00000005573
|
tmem237b
|
transmembrane protein 237b |
| chr8_+_7801060 | 0.40 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr4_+_70151160 | 0.39 |
ENSDART00000111816
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr20_+_15600167 | 0.36 |
ENSDART00000171991
|
faslg
|
Fas ligand (TNF superfamily, member 6) |
| chr2_-_36500347 | 0.36 |
ENSDART00000110528
|
BX901889.1
|
|
| chr23_+_39462208 | 0.34 |
ENSDART00000136707
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr11_-_2838699 | 0.34 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr3_-_36602069 | 0.33 |
ENSDART00000165414
|
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr23_-_36857964 | 0.33 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr19_+_5480327 | 0.31 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr25_-_3058687 | 0.28 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr8_-_12867434 | 0.27 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr4_-_2545310 | 0.26 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr9_+_24008879 | 0.24 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr5_-_16218777 | 0.24 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr17_+_25481466 | 0.20 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr17_+_44697604 | 0.18 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr21_+_8276989 | 0.18 |
ENSDART00000028265
|
nr5a1b
|
nuclear receptor subfamily 5, group A, member 1b |
| chr7_+_7696665 | 0.15 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr18_+_2228737 | 0.14 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr5_+_56023186 | 0.12 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr24_-_36680261 | 0.08 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr3_-_31079186 | 0.07 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr2_+_38373272 | 0.05 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr15_-_43284021 | 0.04 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr8_-_13046089 | 0.04 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb7a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 0.8 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 4.8 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.5 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.8 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 1.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.9 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.3 | GO:0032877 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 3.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 1.3 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 2.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.8 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 1.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 1.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 1.9 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 0.7 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 3.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 1.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.8 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.8 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 4.8 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 3.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.9 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 1.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 5.7 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |