Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
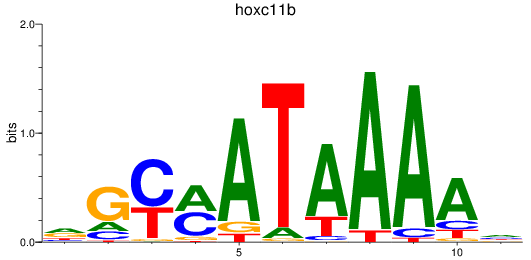
Results for hoxc11b
Z-value: 0.88
Transcription factors associated with hoxc11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc11b
|
ENSDARG00000102631 | homeobox C11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc11b | dr11_v1_chr11_+_2180072_2180072 | 0.46 | 3.2e-06 | Click! |
Activity profile of hoxc11b motif
Sorted Z-values of hoxc11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_29161609 | 16.84 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr10_+_36650222 | 14.08 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr11_-_18253111 | 10.02 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr7_+_39386982 | 9.63 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr8_+_11687254 | 9.50 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr4_+_7391400 | 9.20 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr20_+_40237441 | 8.81 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr14_-_25949951 | 8.66 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr4_+_7391110 | 8.21 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr14_+_15495088 | 7.42 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr7_-_16562200 | 7.21 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr24_+_39105051 | 7.17 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr14_+_15597049 | 7.09 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr14_-_33613794 | 6.93 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr3_+_27027781 | 6.72 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr9_-_41784799 | 6.37 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr25_+_20089986 | 6.31 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr6_-_40697585 | 6.30 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr24_+_5237753 | 6.21 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr12_-_20373058 | 5.99 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr16_+_13993285 | 5.23 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr23_+_18722715 | 5.18 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr7_-_23745984 | 4.90 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr17_+_15788100 | 4.73 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr24_-_29822913 | 4.70 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr5_+_13385837 | 4.49 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr10_+_26612321 | 4.40 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr8_+_47633438 | 4.36 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr22_-_4644484 | 3.91 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr11_+_24313931 | 3.85 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr14_-_15171435 | 3.65 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr2_-_44283554 | 3.48 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr7_+_34290051 | 3.39 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr3_+_52545400 | 3.37 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr14_+_30279391 | 3.26 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr5_-_48307804 | 3.25 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr12_-_25380028 | 3.17 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr23_+_36074798 | 3.16 |
ENSDART00000133760
ENSDART00000103146 |
hoxc11a
|
homeobox C11a |
| chr3_+_52545014 | 3.12 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr15_-_34567370 | 3.09 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr4_+_12292274 | 3.06 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr23_+_18722915 | 3.05 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr7_+_27317174 | 3.04 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr7_-_41420226 | 3.04 |
ENSDART00000074155
|
abcf2b
|
ATP-binding cassette, sub-family F (GCN20), member 2b |
| chr5_-_9073433 | 2.94 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr25_+_26798673 | 2.79 |
ENSDART00000157235
|
ca12
|
carbonic anhydrase XII |
| chr23_+_36122058 | 2.78 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr7_+_46003449 | 2.77 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr8_-_14121634 | 2.75 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr4_-_16836006 | 2.73 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr8_-_25338709 | 2.59 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr19_+_3840955 | 2.56 |
ENSDART00000172305
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr23_+_25354856 | 2.55 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr20_+_52546186 | 2.51 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr2_+_20406399 | 2.49 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr5_-_42272517 | 2.47 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr21_-_20840714 | 2.37 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr5_+_70155935 | 2.35 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr20_+_33532296 | 2.32 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr19_-_14191592 | 2.27 |
ENSDART00000164594
|
tbxta
|
T-box transcription factor Ta |
| chr11_-_25257595 | 2.26 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr21_-_21781158 | 2.20 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr23_+_36052944 | 2.19 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr18_+_14342326 | 2.19 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr19_+_19761966 | 2.13 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr5_-_40510397 | 2.12 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr1_+_961607 | 2.12 |
ENSDART00000184660
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 |
| chr24_+_7782313 | 2.06 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr11_+_25257022 | 2.06 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_+_19747430 | 2.04 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr6_+_30703828 | 2.03 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr6_+_13083146 | 1.98 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr22_+_1911269 | 1.91 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr18_+_910992 | 1.85 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr10_+_22775253 | 1.84 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr11_-_25257045 | 1.83 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr22_-_34551568 | 1.79 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr11_-_25539323 | 1.74 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr12_+_27117609 | 1.73 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr23_+_27068225 | 1.69 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr22_-_14115292 | 1.69 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr22_-_10570749 | 1.67 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr14_+_23184517 | 1.64 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr11_+_42556395 | 1.63 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr11_+_21053488 | 1.61 |
ENSDART00000189860
|
zgc:113307
|
zgc:113307 |
| chr17_+_27456804 | 1.61 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr14_-_3229384 | 1.59 |
ENSDART00000167247
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr23_+_7379728 | 1.58 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr13_-_37465524 | 1.55 |
ENSDART00000100324
ENSDART00000147336 |
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr20_-_33961697 | 1.55 |
ENSDART00000061765
|
selp
|
selectin P |
| chr1_+_8694196 | 1.55 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr6_+_58622831 | 1.55 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr7_+_18075504 | 1.54 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr7_-_58952382 | 1.47 |
ENSDART00000167076
|
mrc1a
|
mannose receptor, C type 1a |
| chr22_-_11648094 | 1.46 |
ENSDART00000191791
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr17_+_8799661 | 1.43 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr24_+_1042594 | 1.42 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr7_+_11543999 | 1.41 |
ENSDART00000173676
|
il16
|
interleukin 16 |
| chr25_-_20258508 | 1.39 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr6_-_6448519 | 1.38 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr13_-_319996 | 1.36 |
ENSDART00000148675
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr2_-_19520324 | 1.33 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr7_-_30280934 | 1.33 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr21_+_5531138 | 1.32 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr22_-_5933844 | 1.31 |
ENSDART00000163370
ENSDART00000189331 |
si:rp71-36a1.2
|
si:rp71-36a1.2 |
| chr18_+_619619 | 1.30 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr24_+_30392834 | 1.28 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr24_-_30091937 | 1.28 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr21_-_39931285 | 1.25 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr10_-_10864331 | 1.25 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr15_+_31344472 | 1.24 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr3_-_53091946 | 1.23 |
ENSDART00000187297
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr21_-_17296789 | 1.22 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr20_-_48701593 | 1.22 |
ENSDART00000132835
|
pax1b
|
paired box 1b |
| chr11_-_38899816 | 1.21 |
ENSDART00000141515
|
pimr127
|
Pim proto-oncogene, serine/threonine kinase, related 127 |
| chr11_-_38928760 | 1.20 |
ENSDART00000146277
|
si:ch211-122l14.6
|
si:ch211-122l14.6 |
| chr19_+_7810028 | 1.20 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr24_-_10014512 | 1.19 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr6_-_46768040 | 1.18 |
ENSDART00000154071
|
igfn1.2
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 2 |
| chr5_-_41307550 | 1.16 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr7_+_9981757 | 1.15 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr4_-_149334 | 1.14 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr7_-_7692723 | 1.14 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr10_+_506538 | 1.13 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr9_-_1949915 | 1.11 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr7_-_18508815 | 1.11 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr23_+_44349252 | 1.09 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr18_-_15559817 | 1.08 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr19_+_19762183 | 1.08 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr8_-_38159805 | 1.07 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr3_+_25154078 | 1.05 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr20_+_46202188 | 1.05 |
ENSDART00000100523
|
taar13c
|
trace amine associated receptor 13c |
| chr2_+_14992879 | 1.05 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr13_+_18520738 | 1.04 |
ENSDART00000113952
|
tlr4al
|
toll-like receptor 4a, like |
| chr3_+_31851004 | 1.04 |
ENSDART00000181422
|
BX324157.1
|
|
| chr18_-_48508585 | 1.03 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr16_-_10547781 | 1.03 |
ENSDART00000166495
|
g6fl
|
g6f-like |
| chr11_-_15090564 | 1.01 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr20_-_26491567 | 1.00 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr2_+_42260021 | 1.00 |
ENSDART00000124702
ENSDART00000140203 ENSDART00000184079 ENSDART00000193349 |
ftr04
|
finTRIM family, member 4 |
| chr8_+_41037541 | 0.99 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr24_+_25258904 | 0.96 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr25_+_16080181 | 0.96 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr11_+_37049105 | 0.96 |
ENSDART00000155408
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr12_+_19356623 | 0.95 |
ENSDART00000078284
|
dmc1
|
DNA meiotic recombinase 1 |
| chr10_-_6427362 | 0.94 |
ENSDART00000166774
|
ca9
|
carbonic anhydrase IX |
| chr6_+_27991943 | 0.94 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr5_-_57723929 | 0.93 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr1_+_42224769 | 0.93 |
ENSDART00000177496
ENSDART00000184778 ENSDART00000110860 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr8_+_12951155 | 0.92 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr1_-_44933094 | 0.92 |
ENSDART00000147527
|
si:dkey-9i23.14
|
si:dkey-9i23.14 |
| chr22_-_4944795 | 0.92 |
ENSDART00000147116
|
pku300
|
pku300 |
| chr22_+_8612462 | 0.92 |
ENSDART00000114586
|
CR450686.1
|
|
| chr15_-_5309213 | 0.91 |
ENSDART00000174081
|
or120-1
|
odorant receptor, family E, subfamily 120, member 1 |
| chr4_-_73136420 | 0.90 |
ENSDART00000160907
|
si:ch73-170d6.3
|
si:ch73-170d6.3 |
| chr15_+_29024895 | 0.89 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr1_+_44439661 | 0.88 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr7_-_44957503 | 0.87 |
ENSDART00000165077
|
cdh16
|
cadherin 16, KSP-cadherin |
| chr10_+_40756352 | 0.87 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr13_+_28417297 | 0.87 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr23_-_36003282 | 0.86 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr12_+_27704015 | 0.86 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr2_+_19522082 | 0.85 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr7_-_55051692 | 0.84 |
ENSDART00000170637
|
tpcn2
|
two pore segment channel 2 |
| chr10_+_40700311 | 0.84 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr13_+_15933168 | 0.83 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr2_-_19520690 | 0.83 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr2_+_19578446 | 0.83 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr12_-_18872717 | 0.81 |
ENSDART00000126300
|
shisa8b
|
shisa family member 8b |
| chr9_-_52490579 | 0.81 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr22_-_11648322 | 0.81 |
ENSDART00000109596
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr10_+_40660772 | 0.79 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr13_+_30169681 | 0.79 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr22_-_31517300 | 0.77 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr13_+_13668991 | 0.77 |
ENSDART00000148266
|
pimr48
|
Pim proto-oncogene, serine/threonine kinase, related 48 |
| chr4_+_64338860 | 0.77 |
ENSDART00000165964
|
si:ch211-223a21.1
|
si:ch211-223a21.1 |
| chr11_-_15090118 | 0.77 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr6_+_52869892 | 0.77 |
ENSDART00000146143
|
si:dkeyp-3f10.16
|
si:dkeyp-3f10.16 |
| chr21_-_20341836 | 0.76 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr11_+_25112269 | 0.75 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr12_+_1000526 | 0.75 |
ENSDART00000182556
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr12_-_33817114 | 0.73 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr12_-_18872927 | 0.73 |
ENSDART00000187717
|
shisa8b
|
shisa family member 8b |
| chr3_-_58798377 | 0.73 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr8_-_28655338 | 0.73 |
ENSDART00000017440
|
mc3r
|
melanocortin 3 receptor |
| chr2_-_19576640 | 0.72 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr2_-_32366287 | 0.72 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr17_+_11808031 | 0.71 |
ENSDART00000190708
|
BX276117.1
|
|
| chr19_+_42469058 | 0.70 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr24_+_26134209 | 0.69 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr4_+_43522945 | 0.69 |
ENSDART00000183921
ENSDART00000181832 |
si:dkeyp-53e4.4
|
si:dkeyp-53e4.4 |
| chr21_+_45819662 | 0.69 |
ENSDART00000193362
ENSDART00000184255 |
pitx1
|
paired-like homeodomain 1 |
| chr9_-_23156908 | 0.68 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr10_-_40484531 | 0.67 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr12_-_11258404 | 0.67 |
ENSDART00000149229
|
si:ch73-30l9.1
|
si:ch73-30l9.1 |
| chr11_-_1392468 | 0.66 |
ENSDART00000004423
|
iars
|
isoleucyl-tRNA synthetase |
| chr11_+_37049347 | 0.66 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr6_+_4528631 | 0.65 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr7_+_38529263 | 0.64 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr9_-_48370645 | 0.64 |
ENSDART00000140185
|
col28a2a
|
collagen, type XXVIII, alpha 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 2.0 | 10.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 1.8 | 7.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 1.8 | 14.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 1.6 | 4.8 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 1.6 | 6.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 1.3 | 6.5 | GO:0071072 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.9 | 5.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.8 | 3.8 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 33.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.6 | 1.8 | GO:1901296 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.6 | 2.3 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.5 | 2.4 | GO:2000051 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.5 | 3.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 1.6 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.4 | 1.6 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.4 | 1.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.3 | 6.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.3 | 1.6 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.3 | 2.9 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 1.5 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.3 | 8.9 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 18.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.3 | 2.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.3 | 2.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.3 | 1.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 1.6 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.3 | 1.3 | GO:0019860 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.2 | 1.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.2 | 0.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 5.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 4.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.2 | 4.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 2.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 0.8 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 2.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 0.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.6 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 4.7 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 1.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 1.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.9 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 2.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.5 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.8 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.1 | 0.3 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 9.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 1.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.5 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 3.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 5.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.4 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 2.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.7 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 4.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 3.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 3.4 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.1 | 0.4 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 1.6 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 3.2 | GO:0060914 | heart formation(GO:0060914) |
| 0.1 | 1.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 3.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.0 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.5 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 5.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.2 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 2.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 3.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.0 | 8.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.2 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 1.0 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 1.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 2.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.8 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 1.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 2.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 1.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 2.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 4.7 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.7 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.0 | 1.3 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.4 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.8 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.9 | 4.7 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 33.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 9.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 2.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 2.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 5.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 2.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 1.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 3.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 0.5 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 2.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 3.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 7.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 7.8 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 6.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 8.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 19.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 1.3 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 24.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 3.1 | 18.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.1 | 6.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 1.7 | 5.2 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.2 | 4.7 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 1.1 | 4.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.0 | 4.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.9 | 6.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.9 | 7.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.6 | 1.7 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.6 | 9.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 1.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 2.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 3.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.3 | 1.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 2.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.3 | 0.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 7.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 1.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 1.3 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.2 | 0.7 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 8.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 6.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.2 | 1.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.2 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 1.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 0.9 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 3.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 2.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.0 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 3.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 2.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.8 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 4.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 3.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 3.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 1.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 2.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 4.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 7.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.7 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 1.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.8 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0071916 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 18.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 4.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 11.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 3.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 14.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 5.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 6.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 8.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 4.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |