Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
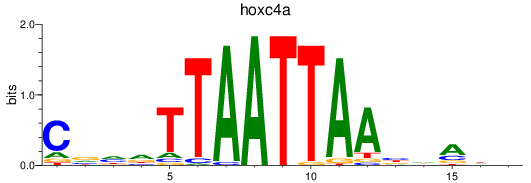
Results for hoxc4a
Z-value: 0.41
Transcription factors associated with hoxc4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc4a
|
ENSDARG00000070338 | homeobox C4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc4a | dr11_v1_chr23_+_36130883_36130883 | -0.04 | 7.3e-01 | Click! |
Activity profile of hoxc4a motif
Sorted Z-values of hoxc4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_46158078 | 5.15 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr16_-_42894628 | 2.60 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr8_-_18537866 | 2.33 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr9_-_22834860 | 2.01 |
ENSDART00000146486
|
neb
|
nebulin |
| chr12_-_47648538 | 1.97 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr16_+_12836143 | 1.84 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr24_-_25691020 | 1.82 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr5_-_38451082 | 1.76 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr6_-_51771634 | 1.31 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr25_-_27621268 | 1.16 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr7_+_4474880 | 1.15 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr14_-_34771371 | 1.13 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr1_+_6172786 | 1.11 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr8_+_30664077 | 1.11 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr8_-_53044300 | 1.11 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr10_-_13343831 | 1.05 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr12_-_19119176 | 1.02 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr3_+_5575313 | 1.01 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr12_+_20641102 | 0.96 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr25_-_13490744 | 0.94 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr9_-_22099536 | 0.92 |
ENSDART00000101923
|
CR391987.1
|
|
| chr22_-_19552796 | 0.88 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr19_+_10855158 | 0.87 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr23_+_4709607 | 0.86 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr1_+_51039558 | 0.85 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr15_-_14552101 | 0.82 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr20_-_43663494 | 0.80 |
ENSDART00000144564
|
BX470188.1
|
|
| chr13_-_12602920 | 0.77 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr3_+_62000822 | 0.77 |
ENSDART00000106680
|
rai1
|
retinoic acid induced 1 |
| chr3_-_59297532 | 0.76 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr18_+_3037998 | 0.73 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr3_-_50443607 | 0.72 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr19_+_5480327 | 0.70 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr10_+_11261576 | 0.70 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr12_+_20641471 | 0.70 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr9_-_1986014 | 0.70 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr18_+_7456597 | 0.65 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr4_+_33017081 | 0.63 |
ENSDART00000151897
|
si:dkey-26h11.1
|
si:dkey-26h11.1 |
| chr20_-_23842631 | 0.63 |
ENSDART00000153079
|
si:dkey-15j16.3
|
si:dkey-15j16.3 |
| chr14_-_858985 | 0.63 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr1_-_513762 | 0.63 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr9_-_12811936 | 0.62 |
ENSDART00000188490
|
myo10l3
|
myosin X-like 3 |
| chr3_-_3798755 | 0.61 |
ENSDART00000170535
|
gucy2c
|
guanylate cyclase 2C |
| chr23_-_21453614 | 0.61 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr2_-_7845110 | 0.61 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr2_+_37140448 | 0.61 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr22_+_1615691 | 0.60 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr4_-_13548806 | 0.59 |
ENSDART00000067155
|
il22
|
interleukin 22 |
| chr17_+_31914877 | 0.59 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr17_-_10122204 | 0.58 |
ENSDART00000160751
|
BX088587.1
|
|
| chr2_+_42318012 | 0.58 |
ENSDART00000138137
|
ftr08
|
finTRIM family, member 8 |
| chr2_-_16216568 | 0.58 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr11_-_2838699 | 0.57 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr2_-_30668580 | 0.57 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr9_+_11281969 | 0.56 |
ENSDART00000110691
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr3_-_13921173 | 0.56 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr19_+_25154066 | 0.55 |
ENSDART00000163220
|
si:ch211-239d6.2
|
si:ch211-239d6.2 |
| chr25_+_13321307 | 0.55 |
ENSDART00000161284
|
si:ch211-194m7.5
|
si:ch211-194m7.5 |
| chr23_-_1660708 | 0.55 |
ENSDART00000175138
|
CU693481.1
|
|
| chr5_+_71802014 | 0.54 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr18_+_7456888 | 0.52 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr10_+_42423318 | 0.52 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr18_+_48168755 | 0.52 |
ENSDART00000128189
|
smtla
|
somatolactin alpha |
| chr6_-_33878665 | 0.51 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr14_-_4556896 | 0.50 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr17_+_2162916 | 0.50 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr13_-_9450210 | 0.49 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr4_-_2219705 | 0.49 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr12_-_26153101 | 0.49 |
ENSDART00000076051
|
opn4b
|
opsin 4b |
| chr3_-_34136778 | 0.48 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr9_+_30211038 | 0.48 |
ENSDART00000190847
|
senp7a
|
SUMO1/sentrin specific peptidase 7a |
| chr22_+_1556948 | 0.48 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr25_-_13320986 | 0.48 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr7_+_38811800 | 0.47 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr1_+_21731382 | 0.47 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr11_+_7214353 | 0.46 |
ENSDART00000156764
|
nwd1
|
NACHT and WD repeat domain containing 1 |
| chr22_+_23359369 | 0.45 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr22_+_1568306 | 0.45 |
ENSDART00000163223
|
si:ch211-255f4.9
|
si:ch211-255f4.9 |
| chr20_+_40457599 | 0.45 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr5_+_33498253 | 0.44 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr4_+_36150332 | 0.44 |
ENSDART00000161697
|
znf1116
|
zinc finger protein 1116 |
| chr5_+_34623107 | 0.43 |
ENSDART00000184126
|
enc1
|
ectodermal-neural cortex 1 |
| chr20_-_9095105 | 0.42 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr5_-_28109416 | 0.42 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr5_-_16218777 | 0.41 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr2_+_38373272 | 0.41 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr1_+_51721851 | 0.41 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr9_+_1139378 | 0.40 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr9_+_48761455 | 0.40 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr6_+_11249706 | 0.40 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr17_-_20558961 | 0.40 |
ENSDART00000155993
|
sh3pxd2ab
|
SH3 and PX domains 2Ab |
| chr20_-_31308805 | 0.40 |
ENSDART00000147045
|
hpcal1
|
hippocalcin-like 1 |
| chr21_+_8276989 | 0.39 |
ENSDART00000028265
|
nr5a1b
|
nuclear receptor subfamily 5, group A, member 1b |
| chr8_+_29742237 | 0.39 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr15_+_31899312 | 0.39 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr1_-_31534089 | 0.38 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr7_-_57509447 | 0.38 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr1_+_418869 | 0.38 |
ENSDART00000152173
|
tpp2
|
tripeptidyl peptidase 2 |
| chr10_-_31015535 | 0.37 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr3_+_23029484 | 0.37 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr23_-_16485190 | 0.37 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr19_+_31873308 | 0.37 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr22_+_737211 | 0.36 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr19_+_3213914 | 0.35 |
ENSDART00000193144
|
zgc:86598
|
zgc:86598 |
| chr2_+_3201345 | 0.34 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr23_-_19225709 | 0.34 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr14_-_451555 | 0.34 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr1_+_9086942 | 0.34 |
ENSDART00000055023
|
cacng3a
|
calcium channel, voltage-dependent, gamma subunit 3a |
| chr24_-_38657683 | 0.33 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr2_+_47708853 | 0.33 |
ENSDART00000124307
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr7_+_21272833 | 0.33 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr11_-_45141309 | 0.32 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr15_-_17619306 | 0.31 |
ENSDART00000184011
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr6_+_612594 | 0.31 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr13_+_29462249 | 0.30 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr15_+_22867174 | 0.30 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr9_+_23895711 | 0.30 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr4_+_77943184 | 0.30 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr1_+_49668423 | 0.30 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr4_+_45148652 | 0.29 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr3_+_40164129 | 0.29 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr6_+_15373153 | 0.29 |
ENSDART00000155865
|
tmtops2a
|
teleost multiple tissue opsin 2a |
| chr10_-_14556978 | 0.29 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr1_+_11882186 | 0.29 |
ENSDART00000193419
|
snx8b
|
sorting nexin 8b |
| chr20_+_32552912 | 0.28 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr17_+_50524573 | 0.28 |
ENSDART00000187974
|
CR382385.1
|
|
| chr12_-_6880694 | 0.28 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr1_-_19502322 | 0.28 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr13_+_12299997 | 0.27 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr2_-_22631498 | 0.27 |
ENSDART00000146425
|
stk25b
|
serine/threonine kinase 25b |
| chr4_+_69191065 | 0.26 |
ENSDART00000170595
|
znf1075
|
zinc finger protein 1075 |
| chr14_-_32486757 | 0.26 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr7_-_52388734 | 0.24 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr23_+_16638639 | 0.24 |
ENSDART00000143545
|
snphb
|
syntaphilin b |
| chr22_+_5532003 | 0.24 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr1_+_57787642 | 0.24 |
ENSDART00000127091
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr6_+_11250033 | 0.24 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr11_+_45436703 | 0.24 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr3_-_48716422 | 0.23 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr9_-_14504834 | 0.23 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr2_+_2223837 | 0.22 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr8_+_7801060 | 0.22 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr18_+_17827149 | 0.22 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr21_+_25802190 | 0.22 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr14_-_413273 | 0.22 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr5_-_21422390 | 0.21 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr17_-_15229787 | 0.20 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr2_+_20605925 | 0.20 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr19_+_19032141 | 0.19 |
ENSDART00000192432
|
cpne4b
|
copine IVb |
| chr24_+_21540842 | 0.19 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr12_+_46742805 | 0.19 |
ENSDART00000192236
|
plaub
|
plasminogen activator, urokinase b |
| chr8_-_12867128 | 0.18 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_+_105748 | 0.18 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr3_+_3681116 | 0.18 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr6_+_11250316 | 0.18 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr12_-_31794908 | 0.17 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr23_-_1348933 | 0.17 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr14_+_32430982 | 0.17 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr3_+_26288981 | 0.16 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr8_-_12867434 | 0.16 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr13_-_35808904 | 0.16 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr15_+_5360407 | 0.15 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr16_-_12173399 | 0.14 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr11_+_34760628 | 0.14 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr22_-_12160283 | 0.14 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr14_-_26425416 | 0.14 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr1_+_513986 | 0.14 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr24_+_24461341 | 0.14 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr23_+_28809002 | 0.14 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr21_-_22635245 | 0.13 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr12_+_17620067 | 0.13 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B |
| chr7_-_66868543 | 0.12 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr24_+_24461558 | 0.12 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr6_+_35052721 | 0.12 |
ENSDART00000191090
ENSDART00000082940 |
uhmk1
|
U2AF homology motif (UHM) kinase 1 |
| chr14_+_22591624 | 0.10 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr24_+_744713 | 0.10 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr12_+_31735159 | 0.09 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr2_+_50608099 | 0.09 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr9_-_27398369 | 0.08 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr12_-_35830625 | 0.08 |
ENSDART00000180028
|
CU459056.1
|
|
| chr23_-_17657348 | 0.07 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr18_+_14619544 | 0.07 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr18_-_5527050 | 0.06 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr24_+_7800486 | 0.05 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr17_+_51682429 | 0.05 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr3_+_36284986 | 0.05 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr7_+_24573721 | 0.04 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr2_-_17044959 | 0.04 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr12_+_7497882 | 0.04 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr22_+_34784075 | 0.03 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr11_+_30817943 | 0.03 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr10_+_26747755 | 0.02 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr19_+_43359075 | 0.02 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr6_+_24420523 | 0.02 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr21_-_13123176 | 0.00 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0034398 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 1.8 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 2.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.3 | 2.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 0.9 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 1.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 0.9 | GO:0097242 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.2 | 2.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 5.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.2 | 0.6 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 2.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.3 | GO:0097052 | ketone catabolic process(GO:0042182) L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.4 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.4 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 0.8 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.6 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.5 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 1.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 | 0.4 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.5 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.2 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 0.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.8 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.1 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.6 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 1.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 2.0 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.2 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 4.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 1.0 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 0.5 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.2 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 1.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 3.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.9 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.4 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.4 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 2.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0019838 | growth factor binding(GO:0019838) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 3.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 5.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |