Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
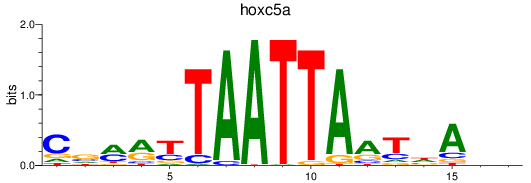
Results for hoxc5a
Z-value: 0.67
Transcription factors associated with hoxc5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc5a
|
ENSDARG00000070340 | homeobox C5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc5a | dr11_v1_chr23_+_36118738_36118738 | -0.01 | 9.3e-01 | Click! |
Activity profile of hoxc5a motif
Sorted Z-values of hoxc5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_17712393 | 12.46 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr15_-_20939579 | 7.82 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr3_-_32818607 | 7.60 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr19_+_46158078 | 7.57 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr3_+_26145013 | 7.41 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr25_+_31267268 | 7.32 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr5_-_71705191 | 6.40 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr8_-_1051438 | 4.73 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr15_-_46779934 | 4.31 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr13_+_9432501 | 4.29 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr9_+_310331 | 4.18 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr5_-_41494831 | 4.18 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr24_+_38301080 | 3.88 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr9_+_307863 | 3.68 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr25_+_14087045 | 3.65 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr19_-_25149598 | 3.58 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr25_+_37293312 | 3.54 |
ENSDART00000086737
ENSDART00000161595 |
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr24_-_25691020 | 3.50 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr19_-_25149034 | 3.40 |
ENSDART00000148432
ENSDART00000175266 |
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr23_+_36460239 | 3.39 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr22_-_10459880 | 3.20 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr12_+_20641471 | 3.18 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr25_+_31227747 | 3.15 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr4_+_77943184 | 3.00 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr8_-_18537866 | 2.97 |
ENSDART00000148802
ENSDART00000148962 ENSDART00000149506 |
nexn
|
nexilin (F actin binding protein) |
| chr5_+_56023186 | 2.95 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr12_+_20641102 | 2.89 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr22_-_31060579 | 2.80 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr16_-_24605969 | 2.80 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr7_+_31891110 | 2.78 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr7_+_59020972 | 2.78 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr12_-_464007 | 2.74 |
ENSDART00000106669
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr25_+_31276842 | 2.72 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr18_-_48547564 | 2.66 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr12_+_48634927 | 2.63 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr19_+_2631565 | 2.59 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr13_-_31622195 | 2.56 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr9_-_42418470 | 2.54 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr7_-_66868543 | 2.52 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr12_-_48188928 | 2.51 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr24_-_40744672 | 2.50 |
ENSDART00000160672
|
CU633479.1
|
|
| chr11_+_705727 | 2.48 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr5_-_63302944 | 2.44 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr10_-_31015535 | 2.42 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr2_+_24304854 | 2.40 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr24_+_19415124 | 2.36 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr19_+_43297546 | 2.34 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr17_-_49978986 | 2.30 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr5_-_37103487 | 2.27 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr1_-_25966068 | 2.25 |
ENSDART00000137869
ENSDART00000134192 |
synpo2b
|
synaptopodin 2b |
| chr23_+_41679586 | 2.17 |
ENSDART00000067662
|
CU914487.1
|
|
| chr25_+_33192404 | 2.14 |
ENSDART00000193592
|
TPM1 (1 of many)
|
zgc:171719 |
| chr2_+_20605925 | 2.09 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr8_+_31717175 | 2.06 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr24_-_33291784 | 2.05 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr3_+_26342768 | 2.03 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr16_+_12836143 | 2.02 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr15_-_12011390 | 1.98 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr15_-_43284021 | 1.97 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr24_+_32176155 | 1.96 |
ENSDART00000003745
|
vim
|
vimentin |
| chr6_-_7020162 | 1.96 |
ENSDART00000148982
|
bin1b
|
bridging integrator 1b |
| chr17_-_20558961 | 1.95 |
ENSDART00000155993
|
sh3pxd2ab
|
SH3 and PX domains 2Ab |
| chr12_+_3078221 | 1.95 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr24_+_35387517 | 1.94 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr15_-_17618800 | 1.94 |
ENSDART00000157185
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr3_+_30922947 | 1.92 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr6_-_40842768 | 1.92 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr17_-_37395460 | 1.91 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr6_-_51771634 | 1.91 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr12_+_22580579 | 1.90 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr3_+_58833306 | 1.90 |
ENSDART00000113223
|
igl1c3
|
immunoglobulin light 1 constant 3 |
| chr16_-_42894628 | 1.88 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr16_+_54209504 | 1.88 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr1_-_25966411 | 1.85 |
ENSDART00000193375
|
synpo2b
|
synaptopodin 2b |
| chr5_+_11840905 | 1.83 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr8_+_31716872 | 1.78 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr23_+_43255328 | 1.77 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr2_+_36004381 | 1.77 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr10_-_22095505 | 1.73 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr3_+_2971852 | 1.72 |
ENSDART00000059271
|
BX004816.1
|
|
| chr24_+_2495197 | 1.72 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr1_-_22851481 | 1.72 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr13_-_35808904 | 1.71 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr3_-_34136778 | 1.71 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr25_+_8356707 | 1.71 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr8_+_34731982 | 1.70 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr22_+_737211 | 1.69 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr22_+_37631234 | 1.62 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr18_+_35128685 | 1.62 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr7_+_20512419 | 1.60 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr8_-_2434282 | 1.57 |
ENSDART00000137262
ENSDART00000134044 |
vdac3
|
voltage-dependent anion channel 3 |
| chr10_-_20445549 | 1.56 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr2_-_21438492 | 1.54 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr15_+_31899312 | 1.53 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr3_-_19368435 | 1.52 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr2_+_38373272 | 1.50 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr18_-_8877077 | 1.50 |
ENSDART00000137266
|
si:dkey-95h12.2
|
si:dkey-95h12.2 |
| chr5_-_9824908 | 1.49 |
ENSDART00000169698
|
zgc:158343
|
zgc:158343 |
| chr12_-_33357655 | 1.47 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr10_-_34916208 | 1.45 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr17_+_28102487 | 1.44 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr10_+_44986419 | 1.43 |
ENSDART00000162566
|
h2afvb
|
H2A histone family, member Vb |
| chr9_-_9415000 | 1.41 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr5_-_38451082 | 1.41 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr22_+_19366866 | 1.39 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr6_-_54179860 | 1.39 |
ENSDART00000164283
|
rps10
|
ribosomal protein S10 |
| chr16_-_12316979 | 1.39 |
ENSDART00000182392
|
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr16_+_29509133 | 1.39 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr5_-_1203455 | 1.37 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr2_+_11205795 | 1.36 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr23_+_4709607 | 1.34 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr4_+_76705830 | 1.34 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr10_-_43771447 | 1.34 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr16_+_13965923 | 1.33 |
ENSDART00000103857
|
zgc:162509
|
zgc:162509 |
| chr22_+_19407531 | 1.33 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr1_-_19502322 | 1.32 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr23_+_4689626 | 1.32 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr21_-_40174647 | 1.30 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr4_+_22480169 | 1.29 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr3_-_15999501 | 1.29 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr19_+_14573998 | 1.28 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr12_-_18578432 | 1.28 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr12_-_18578218 | 1.27 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr24_-_27452488 | 1.27 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr16_+_54780544 | 1.26 |
ENSDART00000126646
|
si:zfos-1192g2.3
|
si:zfos-1192g2.3 |
| chr12_+_22680115 | 1.25 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr22_+_37631034 | 1.24 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr3_+_13559199 | 1.23 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr23_-_42232124 | 1.23 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr21_-_44081540 | 1.23 |
ENSDART00000130833
|
FO704810.1
|
|
| chr16_-_35952789 | 1.22 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr5_-_41831646 | 1.22 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr3_-_34136368 | 1.21 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr14_-_858985 | 1.20 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr15_-_25527580 | 1.20 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr25_+_22320738 | 1.20 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr13_-_37620091 | 1.20 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr2_-_7845110 | 1.20 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr5_-_3927692 | 1.19 |
ENSDART00000146840
ENSDART00000058346 |
c1qbp
|
complement component 1, q subcomponent binding protein |
| chr3_+_5575313 | 1.19 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr13_-_33321541 | 1.18 |
ENSDART00000144196
|
si:dkey-71p21.13
|
si:dkey-71p21.13 |
| chr23_-_42752550 | 1.18 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr19_+_47405867 | 1.18 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr21_-_45363871 | 1.17 |
ENSDART00000075443
ENSDART00000182078 ENSDART00000151106 |
PPP2CA
|
zgc:56064 |
| chr2_+_105748 | 1.17 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr21_+_28478663 | 1.17 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr1_+_27690 | 1.16 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr21_+_13387965 | 1.16 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr24_+_22731228 | 1.14 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr4_-_23759192 | 1.13 |
ENSDART00000014685
ENSDART00000131690 |
cpm
|
carboxypeptidase M |
| chr6_-_7720332 | 1.13 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr9_-_47472998 | 1.13 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr16_-_34212912 | 1.12 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr21_-_35534401 | 1.12 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr1_-_58975098 | 1.12 |
ENSDART00000189899
|
CABZ01114581.1
|
|
| chr2_-_57837838 | 1.12 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr3_+_13848226 | 1.12 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr25_+_16098620 | 1.11 |
ENSDART00000142564
ENSDART00000165598 |
far1
|
fatty acyl CoA reductase 1 |
| chr16_+_23431189 | 1.11 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr10_-_41302841 | 1.10 |
ENSDART00000020297
ENSDART00000160174 ENSDART00000183850 ENSDART00000169493 |
brf2
|
BRF2, RNA polymerase III transcription initiation factor |
| chr21_-_22827548 | 1.10 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr6_-_44402358 | 1.10 |
ENSDART00000193007
ENSDART00000193603 |
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr12_-_30549022 | 1.09 |
ENSDART00000102474
|
zgc:158404
|
zgc:158404 |
| chr2_+_9990491 | 1.09 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr13_+_48358467 | 1.08 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr2_+_10878406 | 1.08 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr21_-_32060993 | 1.08 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr16_-_38118003 | 1.08 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr8_-_49728590 | 1.08 |
ENSDART00000135714
ENSDART00000138810 ENSDART00000098319 |
gkap1
|
G kinase anchoring protein 1 |
| chr12_+_23912074 | 1.06 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr25_+_16116740 | 1.06 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr12_-_25294096 | 1.06 |
ENSDART00000183398
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr22_+_11775269 | 1.05 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr7_-_51368681 | 1.04 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr22_-_22301929 | 1.04 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr15_+_23722620 | 1.03 |
ENSDART00000011447
|
sae1
|
SUMO1 activating enzyme subunit 1 |
| chr11_+_45436703 | 1.03 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr12_-_4243268 | 1.02 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr1_+_22851261 | 1.02 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr23_-_42752387 | 1.02 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr19_+_43359075 | 1.02 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr22_-_16042243 | 1.01 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr1_+_51039558 | 1.00 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr20_-_182841 | 0.99 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr21_-_45382112 | 0.99 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr6_+_12527725 | 0.97 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr21_-_43015383 | 0.97 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr23_-_19225709 | 0.96 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr24_+_16985181 | 0.96 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr21_-_1644414 | 0.96 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr5_+_6672870 | 0.95 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr10_+_44719697 | 0.95 |
ENSDART00000158087
|
scarb1
|
scavenger receptor class B, member 1 |
| chr7_+_51774502 | 0.95 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr12_-_47648538 | 0.94 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr14_+_15257658 | 0.94 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr9_-_14504834 | 0.94 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr5_+_1624359 | 0.93 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr7_-_67248829 | 0.93 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr15_+_9327252 | 0.93 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr14_-_24761132 | 0.93 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.4 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 1.6 | 7.9 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.9 | 2.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.8 | 3.8 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.7 | 4.8 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.6 | 2.6 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.6 | 1.9 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.6 | 3.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.6 | 2.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.4 | 2.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 4.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 | 1.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.4 | 2.5 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 2.0 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.4 | 1.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.4 | 3.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.4 | 1.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.4 | 1.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.4 | 1.1 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 1.4 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 2.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 13.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 1.0 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 1.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.3 | 0.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 1.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 | 3.0 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.3 | 1.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.3 | 0.9 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.3 | 0.8 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 7.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.3 | 1.9 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) |
| 0.2 | 1.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 0.7 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 0.9 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 1.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 6.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.2 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.2 | 1.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.2 | 2.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 5.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.5 | GO:0034138 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.2 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.5 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 3.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.6 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 0.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 4.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 4.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.4 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.4 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 1.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.7 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.7 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 2.7 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 0.5 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 1.0 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 1.0 | GO:0072425 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 1.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.6 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 0.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.1 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 0.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 4.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.8 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 2.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.6 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 1.0 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.8 | GO:0007589 | body fluid secretion(GO:0007589) |
| 0.1 | 0.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.6 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.4 | GO:0048855 | post-embryonic foregut morphogenesis(GO:0048618) adenohypophysis morphogenesis(GO:0048855) |
| 0.1 | 1.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 1.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 2.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.6 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 2.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.4 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 1.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 4.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 2.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.2 | GO:0048713 | positive regulation of glial cell differentiation(GO:0045687) regulation of oligodendrocyte differentiation(GO:0048713) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 1.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.4 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 1.1 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.8 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 8.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 1.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 3.4 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 0.6 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 2.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.1 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 1.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.6 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.8 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 1.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 2.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 2.9 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 0.3 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) positive regulation of triglyceride metabolic process(GO:0090208) |
| 0.1 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 3.5 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 1.9 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 3.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 3.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.3 | GO:0009217 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 1.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.7 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 2.1 | GO:1990573 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.4 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 1.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 2.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 1.5 | GO:0016571 | histone methylation(GO:0016571) |
| 0.0 | 1.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.3 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.2 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.2 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 1.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 | 0.5 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 2.5 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 4.4 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.1 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.5 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 1.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.5 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.5 | GO:0042176 | regulation of protein catabolic process(GO:0042176) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.1 | GO:0010821 | regulation of mitochondrion organization(GO:0010821) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.0 | 2.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.4 | GO:0031673 | H zone(GO:0031673) |
| 1.0 | 2.9 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.6 | 9.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.5 | 1.5 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.5 | 1.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 2.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 4.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.4 | 1.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.4 | 1.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.3 | 13.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 4.7 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 1.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 1.2 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 1.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 1.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 2.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 0.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 1.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 1.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 1.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 2.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 3.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.9 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 7.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.9 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.2 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 4.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 2.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 3.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 11.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 3.2 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 2.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 1.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 4.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 2.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 6.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.8 | 2.5 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.8 | 3.8 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.7 | 2.8 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 2.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.4 | 7.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 1.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 4.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 1.1 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.4 | 3.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.3 | 1.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.3 | 2.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.3 | 2.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 2.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 1.5 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.3 | 0.9 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.3 | 2.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 0.9 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 2.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 2.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 4.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 4.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 0.8 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 3.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 3.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 3.2 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.2 | 3.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 0.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 2.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 1.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 2.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 2.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 1.1 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 1.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 4.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 4.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 1.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 2.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.5 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.3 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.6 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 9.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.7 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.1 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.4 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 2.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.5 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 2.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.1 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.4 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 5.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.7 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 4.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 7.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 2.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 7.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 6.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 3.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 8.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 3.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 6.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 3.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.4 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 4.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.8 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |