Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
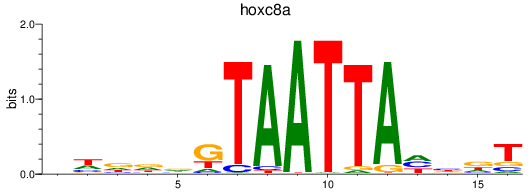
Results for hoxc8a
Z-value: 0.55
Transcription factors associated with hoxc8a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc8a
|
ENSDARG00000070346 | homeobox C8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc8a | dr11_v1_chr23_+_36101185_36101185 | -0.19 | 6.4e-02 | Click! |
Activity profile of hoxc8a motif
Sorted Z-values of hoxc8a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_6188413 | 5.47 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr16_-_44945224 | 4.37 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr13_+_13693722 | 4.18 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr8_+_31717175 | 3.56 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr17_+_23298928 | 3.29 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr8_+_31716872 | 2.88 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr2_-_33687214 | 2.76 |
ENSDART00000147439
|
atp6v0b
|
ATPase H+ transporting V0 subunit b |
| chr16_+_51180938 | 2.73 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr14_-_27121854 | 2.65 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr10_+_42358426 | 2.53 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr8_+_22516728 | 2.45 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr15_-_15357178 | 2.39 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr8_+_18624658 | 2.33 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr2_+_31833997 | 2.30 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr3_+_46628885 | 2.26 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr1_+_52392511 | 2.21 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr2_+_50608099 | 2.19 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr18_+_924949 | 2.18 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr7_-_48667056 | 2.17 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr22_-_1079773 | 2.16 |
ENSDART00000136668
|
si:ch1073-15f12.3
|
si:ch1073-15f12.3 |
| chr1_+_53842344 | 2.14 |
ENSDART00000114506
|
crfb17
|
cytokine receptor family member B17 |
| chr3_+_22442445 | 2.13 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr18_+_7283283 | 2.10 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr8_-_31107537 | 2.09 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr8_+_52637507 | 2.00 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr3_-_31254379 | 1.93 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr11_+_11267493 | 1.89 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr2_-_16380283 | 1.87 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr21_+_30355767 | 1.81 |
ENSDART00000189948
|
CR749164.1
|
|
| chr20_-_34750045 | 1.80 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr16_+_17389116 | 1.79 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr15_+_841383 | 1.78 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr20_+_40457599 | 1.74 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr4_+_76926059 | 1.72 |
ENSDART00000136192
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr1_-_58036509 | 1.65 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr13_+_12299997 | 1.64 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr20_-_7072487 | 1.63 |
ENSDART00000145954
|
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr17_+_26611929 | 1.63 |
ENSDART00000166450
ENSDART00000087023 |
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr21_+_22738939 | 1.62 |
ENSDART00000151342
ENSDART00000079145 |
arhgap42a
|
Rho GTPase activating protein 42a |
| chr6_-_43283122 | 1.59 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr21_-_5856050 | 1.57 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr16_+_13818743 | 1.56 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr15_-_43284021 | 1.53 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr18_+_27738349 | 1.51 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr13_-_35808904 | 1.50 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr11_+_37250839 | 1.46 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr14_-_16082806 | 1.44 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr13_+_38521152 | 1.41 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr6_-_30210378 | 1.37 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr5_+_26213874 | 1.37 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr13_-_31812394 | 1.37 |
ENSDART00000124445
|
sertad4
|
SERTA domain containing 4 |
| chr20_-_9462433 | 1.36 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr4_-_22472653 | 1.35 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr6_-_40713183 | 1.34 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr4_-_815871 | 1.33 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr21_-_35419486 | 1.33 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr20_-_45812144 | 1.31 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr15_+_15856178 | 1.31 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr14_-_47248784 | 1.30 |
ENSDART00000135479
|
fstl5
|
follistatin-like 5 |
| chr2_+_20793982 | 1.26 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr15_-_35252522 | 1.20 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr11_-_21528056 | 1.18 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr6_+_49723289 | 1.16 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr22_-_17474583 | 1.16 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr18_+_17827149 | 1.15 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr20_-_34750363 | 1.09 |
ENSDART00000152845
|
znf395b
|
zinc finger protein 395b |
| chr5_+_30635309 | 1.08 |
ENSDART00000183769
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr4_+_14727018 | 1.07 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr15_-_23420079 | 1.05 |
ENSDART00000104525
ENSDART00000148840 |
kmt2a
|
lysine (K)-specific methyltransferase 2A |
| chr7_-_30174882 | 1.05 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr15_-_37104165 | 1.03 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr1_+_513986 | 1.02 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr3_-_34084387 | 0.97 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr4_+_5333988 | 0.97 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr7_-_46777876 | 0.96 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr24_+_36204028 | 0.94 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr8_-_7567815 | 0.94 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr22_+_29113796 | 0.90 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr21_+_8427059 | 0.80 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr21_-_32097908 | 0.80 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr10_-_35108683 | 0.80 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr13_+_36622100 | 0.79 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr15_+_29140126 | 0.78 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr11_-_40728380 | 0.76 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr23_-_29505645 | 0.75 |
ENSDART00000146458
|
kif1b
|
kinesin family member 1B |
| chr16_-_13623659 | 0.75 |
ENSDART00000168978
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr7_-_32895668 | 0.75 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr22_-_17474781 | 0.75 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr12_-_4532066 | 0.75 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr23_+_579893 | 0.71 |
ENSDART00000189098
|
BX323461.1
|
|
| chr19_+_16064439 | 0.71 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr19_-_33850201 | 0.70 |
ENSDART00000168583
|
otud6b
|
OTU domain containing 6B |
| chr19_-_205104 | 0.70 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr6_-_37744430 | 0.68 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr13_+_31070181 | 0.64 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr23_+_40604951 | 0.64 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr21_-_34032650 | 0.63 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr23_-_35790235 | 0.62 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr15_+_857148 | 0.62 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr24_+_22485710 | 0.61 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr13_+_31321297 | 0.61 |
ENSDART00000143308
|
antxr1d
|
anthrax toxin receptor 1d |
| chr4_+_14727212 | 0.61 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr25_-_7705487 | 0.61 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr13_+_35339182 | 0.59 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr16_-_13623928 | 0.58 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr11_+_30244356 | 0.57 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr4_-_46915962 | 0.55 |
ENSDART00000169555
|
si:ch211-134c10.1
|
si:ch211-134c10.1 |
| chr16_-_28878080 | 0.55 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr6_+_18321627 | 0.54 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr16_+_4497302 | 0.53 |
ENSDART00000081826
ENSDART00000148096 |
ttc29
|
tetratricopeptide repeat domain 29 |
| chr5_+_36768674 | 0.51 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr10_-_21955231 | 0.51 |
ENSDART00000183695
|
FO744833.2
|
|
| chr24_+_26997798 | 0.49 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr20_-_27225876 | 0.49 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr10_+_15224660 | 0.48 |
ENSDART00000137932
|
sh3bp2
|
SH3-domain binding protein 2 |
| chr3_+_30500968 | 0.48 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr21_+_29077509 | 0.48 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr17_-_43556208 | 0.46 |
ENSDART00000188424
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr13_-_46200240 | 0.46 |
ENSDART00000056984
|
ftr69
|
finTRIM family, member 69 |
| chr20_-_29532939 | 0.44 |
ENSDART00000049224
ENSDART00000062377 |
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr19_+_10592778 | 0.44 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr25_-_13490744 | 0.42 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr12_+_18744610 | 0.42 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr24_-_38657683 | 0.39 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr21_-_26490186 | 0.38 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr11_-_44979281 | 0.38 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chrM_+_9052 | 0.37 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr1_+_58628165 | 0.35 |
ENSDART00000158654
|
si:ch73-221f6.4
|
si:ch73-221f6.4 |
| chr11_+_33818179 | 0.34 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr20_+_41021054 | 0.30 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr22_-_6229275 | 0.30 |
ENSDART00000146045
ENSDART00000179730 |
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr7_-_45999333 | 0.28 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr6_+_19948043 | 0.27 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr20_-_14925281 | 0.27 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr1_+_56755536 | 0.25 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr10_+_2899108 | 0.24 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr5_+_27137473 | 0.23 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr15_-_14552101 | 0.23 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr23_-_16485190 | 0.23 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr10_-_26744131 | 0.23 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr11_-_1509773 | 0.19 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr8_-_11004726 | 0.18 |
ENSDART00000192594
ENSDART00000020116 |
trim33
|
tripartite motif containing 33 |
| chr15_+_31899312 | 0.17 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr20_-_14924858 | 0.17 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr16_+_39159752 | 0.15 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr24_+_12894282 | 0.15 |
ENSDART00000061301
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr6_+_49722970 | 0.14 |
ENSDART00000155934
ENSDART00000154738 |
stx16
|
syntaxin 16 |
| chr24_-_7321928 | 0.14 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr20_+_11731039 | 0.11 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr10_+_11355841 | 0.10 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr2_-_36500347 | 0.10 |
ENSDART00000110528
|
BX901889.1
|
|
| chr11_+_45110865 | 0.08 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr15_-_42736433 | 0.06 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr17_-_43556415 | 0.05 |
ENSDART00000190102
ENSDART00000193156 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr16_-_30903930 | 0.05 |
ENSDART00000143996
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr4_+_22343093 | 0.05 |
ENSDART00000023588
|
guca1a
|
guanylate cyclase activator 1A |
| chr2_-_30668580 | 0.03 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr25_+_31868268 | 0.02 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr3_+_7808459 | 0.02 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chrM_+_9735 | 0.01 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr8_-_30204650 | 0.00 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr2_+_16482338 | 0.00 |
ENSDART00000143912
|
fbxo36b
|
F-box protein 36b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc8a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 2.5 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 2.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.3 | 1.9 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.3 | 0.9 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 2.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 1.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.3 | 0.8 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.3 | 0.8 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.2 | 1.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 1.6 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 1.7 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 1.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 5.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.3 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 2.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 1.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 1.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 1.0 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.5 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 3.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.0 | 2.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 2.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 7.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 3.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 2.6 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.9 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 4.2 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.5 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0044218 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 2.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 2.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 1.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 4.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.2 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.9 | 4.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 2.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.4 | 2.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 0.9 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 2.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.0 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 1.7 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.4 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 2.1 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 2.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 5.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 2.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 2.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 3.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 1.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 4.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) ion gated channel activity(GO:0022839) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |