Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
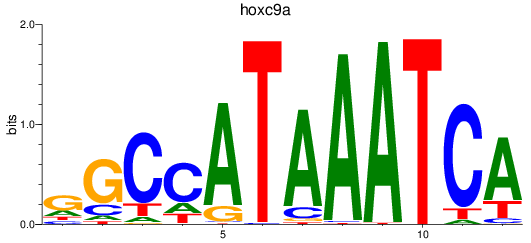
Results for hoxc9a
Z-value: 1.10
Transcription factors associated with hoxc9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc9a
|
ENSDARG00000092809 | homeobox C9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc9a | dr11_v1_chr23_+_36095260_36095260 | 0.07 | 5.3e-01 | Click! |
Activity profile of hoxc9a motif
Sorted Z-values of hoxc9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_47633438 | 26.97 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr17_+_23300827 | 23.70 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr25_-_13842618 | 17.77 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr8_+_16004154 | 17.56 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr11_-_6188413 | 16.71 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr8_+_22931427 | 14.54 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr13_-_43599898 | 12.28 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr3_-_61205711 | 10.95 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr17_+_19626479 | 10.82 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr15_+_8043751 | 10.79 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr10_-_19801821 | 10.78 |
ENSDART00000148013
|
gfra2b
|
GDNF family receptor alpha 2b |
| chr17_+_45454943 | 10.69 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr5_-_51619262 | 10.64 |
ENSDART00000134606
ENSDART00000081249 |
otpb
|
orthopedia homeobox b |
| chr15_-_5467477 | 9.93 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr3_+_34821327 | 9.81 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr1_-_23157583 | 9.80 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr11_-_13341483 | 9.73 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr10_+_26571174 | 9.48 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr2_+_34967022 | 9.48 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr14_+_21783400 | 9.17 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr10_+_20128267 | 9.05 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr25_-_7999756 | 8.21 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr7_-_26408472 | 7.37 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr17_+_18117029 | 7.31 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr9_-_3671911 | 7.24 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr1_+_16144615 | 7.13 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr4_+_10017049 | 7.10 |
ENSDART00000144175
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr4_+_11439511 | 7.07 |
ENSDART00000150485
|
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr17_+_32343121 | 6.83 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr5_-_11573490 | 6.69 |
ENSDART00000109577
|
FO704871.1
|
|
| chr25_+_3058924 | 6.61 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr15_+_22722684 | 6.46 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr18_-_2433011 | 6.38 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr7_-_41964877 | 6.32 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr22_-_21046654 | 6.27 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr4_-_4261673 | 6.27 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr1_-_39943596 | 6.16 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr20_-_36107422 | 6.13 |
ENSDART00000146055
|
ptchd4
|
patched domain containing 4 |
| chr18_-_26510545 | 6.07 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr2_+_2223837 | 6.00 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr11_-_13341051 | 5.21 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr7_+_31051213 | 5.20 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr14_+_11762991 | 5.16 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr19_-_36234185 | 4.93 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr5_-_51619742 | 4.83 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr15_-_14375452 | 4.76 |
ENSDART00000160675
ENSDART00000164028 ENSDART00000171642 |
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr7_-_31938938 | 4.75 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr17_-_20849879 | 4.69 |
ENSDART00000088100
ENSDART00000149630 |
ank3b
|
ankyrin 3b |
| chr9_+_44722205 | 4.50 |
ENSDART00000086176
ENSDART00000145271 ENSDART00000132696 |
nckap1
|
NCK-associated protein 1 |
| chr21_+_42930558 | 4.49 |
ENSDART00000135234
|
stk32a
|
serine/threonine kinase 32A |
| chr2_-_18830722 | 4.48 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr3_-_37699992 | 4.34 |
ENSDART00000151193
|
gpatch8
|
G patch domain containing 8 |
| chr11_+_25044082 | 4.32 |
ENSDART00000123263
|
phf20a
|
PHD finger protein 20, a |
| chr7_+_31051603 | 4.28 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr4_-_20235904 | 4.23 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr7_+_38811800 | 4.17 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr11_-_3334248 | 4.08 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr11_+_16040680 | 3.97 |
ENSDART00000157703
|
agtrap
|
angiotensin II receptor-associated protein |
| chr15_+_28116129 | 3.93 |
ENSDART00000185157
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr1_-_39909985 | 3.78 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr18_-_39288894 | 3.77 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr19_+_41006975 | 3.43 |
ENSDART00000138555
ENSDART00000049842 |
casd1
|
CAS1 domain containing 1 |
| chr15_+_8767650 | 3.36 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr2_-_41723165 | 3.24 |
ENSDART00000155577
|
zgc:110158
|
zgc:110158 |
| chr6_+_38845697 | 3.23 |
ENSDART00000053187
|
stk35l
|
serine/threonine kinase 35, like |
| chr13_+_22316746 | 3.21 |
ENSDART00000188968
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr14_-_4044545 | 3.16 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr19_+_4916233 | 3.06 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr5_-_37871526 | 3.00 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr23_+_20669149 | 2.98 |
ENSDART00000138936
|
arhgef3l
|
Rho guanine nucleotide exchange factor (GEF) 3, like |
| chr17_+_26208630 | 2.95 |
ENSDART00000087084
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr23_-_3721444 | 2.84 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr9_-_25255490 | 2.78 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr2_-_41723487 | 2.68 |
ENSDART00000170171
|
zgc:110158
|
zgc:110158 |
| chr14_+_36231126 | 2.64 |
ENSDART00000141766
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr12_+_27213733 | 2.50 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr14_-_30141162 | 2.47 |
ENSDART00000053916
|
mtnr1ab
|
melatonin receptor 1A b |
| chr21_+_22878991 | 2.43 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr6_+_55032439 | 2.42 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr21_+_22878834 | 2.06 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr20_+_27464721 | 2.05 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr4_+_21867522 | 2.02 |
ENSDART00000140400
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr22_+_10713713 | 1.89 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr25_-_5963535 | 1.86 |
ENSDART00000155751
|
nuak1b
|
NUAK family, SNF1-like kinase, 1b |
| chr9_-_34937025 | 1.76 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr6_+_52804267 | 1.75 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr15_-_14589677 | 1.67 |
ENSDART00000172195
|
coq8b
|
coenzyme Q8B |
| chr5_+_63857055 | 1.64 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr2_-_9259283 | 1.62 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr7_-_41693004 | 1.55 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr20_+_23658791 | 1.51 |
ENSDART00000192145
|
si:ch211-191d2.2
|
si:ch211-191d2.2 |
| chr17_-_51262430 | 1.49 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr22_-_21046843 | 1.41 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr20_-_53949798 | 1.32 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr19_-_27830818 | 1.30 |
ENSDART00000131767
|
papd7
|
PAP associated domain containing 7 |
| chr3_-_40232615 | 1.24 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr3_-_12930217 | 1.22 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr14_-_17072736 | 1.07 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr12_-_20362041 | 1.03 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr12_-_16380961 | 0.89 |
ENSDART00000086984
|
hectd2
|
HECT domain containing 2 |
| chr12_-_16380593 | 0.86 |
ENSDART00000152133
|
hectd2
|
HECT domain containing 2 |
| chr11_+_18873113 | 0.76 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr16_+_37891735 | 0.74 |
ENSDART00000178753
ENSDART00000142104 |
cep162
|
centrosomal protein 162 |
| chr17_-_30839338 | 0.74 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr17_-_25630822 | 0.68 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr21_-_131236 | 0.67 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr9_+_16449398 | 0.54 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr3_-_23461954 | 0.54 |
ENSDART00000040065
|
casc3
|
cancer susceptibility candidate 3 |
| chr3_+_25825043 | 0.38 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr6_-_46875310 | 0.35 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr4_-_18635005 | 0.30 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr22_+_35089031 | 0.23 |
ENSDART00000076040
|
srfa
|
serum response factor a |
| chr3_-_23470986 | 0.21 |
ENSDART00000113135
|
msl1a
|
male-specific lethal 1 homolog a (Drosophila) |
| chr13_+_15004398 | 0.20 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr12_+_18744610 | 0.07 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr3_-_8388344 | 0.07 |
ENSDART00000146856
|
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr23_+_3721042 | 0.06 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr10_+_13000669 | 0.04 |
ENSDART00000158919
ENSDART00000172625 |
lpar1
|
lysophosphatidic acid receptor 1 |
| chr12_-_16380242 | 0.03 |
ENSDART00000168105
|
hectd2
|
HECT domain containing 2 |
| chr4_-_20118468 | 0.03 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.5 | GO:0021767 | mammillary body development(GO:0021767) |
| 2.5 | 9.9 | GO:1900120 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.9 | 6.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.9 | 6.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.9 | 21.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.9 | 9.5 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.9 | 9.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.7 | 4.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.7 | 7.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.6 | 4.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.6 | 2.8 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.5 | 7.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.5 | 9.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.5 | 3.9 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.4 | 10.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 7.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.4 | 3.4 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.4 | 4.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.4 | 16.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.4 | 6.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.4 | 9.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 36.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.3 | 17.8 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.3 | 1.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 9.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.2 | 0.5 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 9.0 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.1 | 1.8 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 27.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 2.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 3.2 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 7.3 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.1 | 1.7 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 6.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 2.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 3.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 10.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 4.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 23.7 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 4.5 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 17.2 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 4.1 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 10.6 | GO:0009968 | negative regulation of signal transduction(GO:0009968) negative regulation of signaling(GO:0023057) |
| 0.0 | 0.2 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 3.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 17.0 | GO:0007267 | cell-cell signaling(GO:0007267) |
| 0.0 | 2.5 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 3.1 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.3 | GO:0071375 | regulation of blood pressure(GO:0008217) cellular response to peptide hormone stimulus(GO:0071375) cellular response to peptide(GO:1901653) |
| 0.0 | 0.7 | GO:0060393 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 33.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 2.0 | 9.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.7 | 4.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.7 | 7.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.5 | 7.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 17.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.4 | 3.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 3.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 4.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.2 | 12.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 4.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 4.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 14.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 9.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.5 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 2.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 9.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 6.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 12.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 10.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 9.9 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 17.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 33.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 3.6 | 10.7 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.9 | 9.5 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.8 | 7.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.8 | 9.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.6 | 2.8 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.6 | 6.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.4 | 2.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 2.0 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.4 | 7.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.4 | 16.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 4.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.4 | 14.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 3.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 23.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.2 | 1.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 4.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 8.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 13.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 3.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.0 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 3.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 9.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 2.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 6.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 13.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 9.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 2.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 5.0 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 3.2 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 4.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 5.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 4.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.4 | 9.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 3.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 2.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 3.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 7.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |