Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
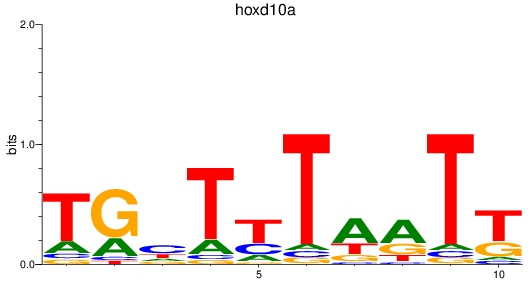
Results for hoxd10a
Z-value: 0.46
Transcription factors associated with hoxd10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd10a
|
ENSDARG00000057859 | homeobox D10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd10a | dr11_v1_chr9_-_1970071_1970071 | -0.36 | 3.9e-04 | Click! |
Activity profile of hoxd10a motif
Sorted Z-values of hoxd10a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45178430 | 3.55 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr14_+_38786298 | 2.99 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr8_+_36142734 | 2.81 |
ENSDART00000159361
ENSDART00000161194 |
mhc2b
|
major histocompatibility complex class II integral membrane beta chain gene |
| chr22_-_38819603 | 2.65 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr17_+_42274825 | 2.03 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr4_-_16824556 | 1.98 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr5_+_22459087 | 1.82 |
ENSDART00000134781
|
BX546499.1
|
|
| chr13_-_31017960 | 1.81 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr5_-_23805387 | 1.76 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr2_-_1486023 | 1.65 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr18_+_15644559 | 1.62 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr5_+_35458346 | 1.59 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr2_+_10280645 | 1.47 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr14_+_26439227 | 1.46 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr11_+_37265692 | 1.45 |
ENSDART00000184691
|
il17rc
|
interleukin 17 receptor C |
| chr13_-_50108337 | 1.45 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr16_-_42186093 | 1.38 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr2_-_42628028 | 1.36 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr5_+_50912729 | 1.27 |
ENSDART00000190837
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr18_+_31056645 | 1.21 |
ENSDART00000159316
|
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr21_+_15295685 | 1.18 |
ENSDART00000135603
|
BX901878.1
|
|
| chr21_-_3796461 | 1.16 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr6_+_29386728 | 1.15 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr25_-_3217115 | 1.15 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr18_+_7363242 | 1.10 |
ENSDART00000133375
|
si:dkey-30c15.17
|
si:dkey-30c15.17 |
| chr20_+_13894123 | 1.04 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr20_+_18993452 | 1.03 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr1_-_52431220 | 1.03 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr6_-_15757867 | 1.02 |
ENSDART00000063665
|
ackr3b
|
atypical chemokine receptor 3b |
| chr18_+_7543347 | 1.01 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr13_+_27232848 | 0.99 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr18_-_20458412 | 0.98 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr20_-_30931139 | 0.98 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr6_-_39521832 | 0.96 |
ENSDART00000065038
|
atf1
|
activating transcription factor 1 |
| chr16_+_12281032 | 0.94 |
ENSDART00000138638
|
si:dkey-26c10.5
|
si:dkey-26c10.5 |
| chr16_-_43233509 | 0.89 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr19_-_3056235 | 0.89 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr10_-_3138403 | 0.88 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr7_-_22981796 | 0.85 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr11_-_39118882 | 0.83 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr23_+_28322986 | 0.83 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr3_-_40768548 | 0.83 |
ENSDART00000004923
|
smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr21_+_21279159 | 0.83 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr3_-_26184018 | 0.83 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr9_+_21151138 | 0.80 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr10_+_44692272 | 0.80 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr10_-_22491353 | 0.79 |
ENSDART00000180783
ENSDART00000159564 |
epob
|
erythropoietin b |
| chr19_+_47299212 | 0.78 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr4_-_76270779 | 0.75 |
ENSDART00000183709
ENSDART00000192689 |
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr21_+_3796620 | 0.70 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr11_-_31039533 | 0.70 |
ENSDART00000127355
|
ier2b
|
immediate early response 2b |
| chr11_-_42752884 | 0.69 |
ENSDART00000186025
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr18_-_2222128 | 0.67 |
ENSDART00000171402
|
pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr7_+_22982201 | 0.66 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr21_-_41025340 | 0.66 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr1_-_37377509 | 0.65 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr11_+_14104417 | 0.64 |
ENSDART00000059752
ENSDART00000186575 |
med16
|
mediator complex subunit 16 |
| chr4_-_67589158 | 0.63 |
ENSDART00000184361
|
BX088712.2
|
|
| chr1_-_669717 | 0.63 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr14_-_32959851 | 0.62 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr3_-_29899172 | 0.60 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr6_-_25165693 | 0.60 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr3_-_11828206 | 0.59 |
ENSDART00000018159
|
si:ch211-262e15.1
|
si:ch211-262e15.1 |
| chr9_-_19489264 | 0.59 |
ENSDART00000122894
|
wdr4
|
WD repeat domain 4 |
| chr8_-_15292197 | 0.59 |
ENSDART00000140867
|
spata6
|
spermatogenesis associated 6 |
| chr6_+_12316438 | 0.59 |
ENSDART00000156854
|
si:dkey-276j7.2
|
si:dkey-276j7.2 |
| chr19_-_35450661 | 0.57 |
ENSDART00000113574
ENSDART00000136895 |
anln
|
anillin, actin binding protein |
| chr7_-_24373662 | 0.56 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr21_+_3796196 | 0.55 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr21_+_5192016 | 0.54 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr19_-_10324573 | 0.54 |
ENSDART00000171795
|
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr5_+_25317061 | 0.54 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr25_-_12412704 | 0.53 |
ENSDART00000168275
|
det1
|
DET1, COP1 ubiquitin ligase partner |
| chr19_+_13994563 | 0.52 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr12_+_30788912 | 0.51 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr11_-_27953135 | 0.50 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr19_+_4051535 | 0.49 |
ENSDART00000167084
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr10_-_26985231 | 0.49 |
ENSDART00000050585
|
znhit2
|
zinc finger, HIT-type containing 2 |
| chr6_+_9793791 | 0.47 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr16_-_26296477 | 0.47 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr9_-_3519717 | 0.46 |
ENSDART00000145043
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr13_+_27232694 | 0.44 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr10_+_22527715 | 0.43 |
ENSDART00000134864
|
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr1_-_18811517 | 0.41 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr1_-_19764038 | 0.38 |
ENSDART00000005733
|
tma16
|
translation machinery associated 16 homolog |
| chr24_+_17069420 | 0.37 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr6_+_1787160 | 0.34 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr7_+_4194306 | 0.33 |
ENSDART00000190454
|
si:dkey-28d5.5
|
si:dkey-28d5.5 |
| chr5_+_6672870 | 0.32 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr9_+_45605410 | 0.32 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr22_+_22302614 | 0.32 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr9_+_3519191 | 0.31 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr13_-_24669258 | 0.30 |
ENSDART00000000831
|
znf511
|
zinc finger protein 511 |
| chr10_-_17484762 | 0.30 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr16_+_41060161 | 0.29 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr14_+_20941038 | 0.29 |
ENSDART00000182539
|
zgc:66433
|
zgc:66433 |
| chr17_-_28707898 | 0.29 |
ENSDART00000135752
ENSDART00000061853 |
ap4s1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr4_+_64339299 | 0.28 |
ENSDART00000191590
|
si:ch211-223a21.1
|
si:ch211-223a21.1 |
| chr10_-_20453995 | 0.28 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr3_-_26191960 | 0.27 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr10_+_41986685 | 0.27 |
ENSDART00000086165
ENSDART00000171983 |
setd1ba
|
SET domain containing 1B, a |
| chr24_+_15655233 | 0.27 |
ENSDART00000143160
|
fbxo15
|
F-box protein 15 |
| chr1_+_32341734 | 0.23 |
ENSDART00000135078
|
pudp
|
pseudouridine 5'-phosphatase |
| chr3_-_17871846 | 0.23 |
ENSDART00000074478
ENSDART00000187941 |
nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr17_+_31621788 | 0.22 |
ENSDART00000111629
ENSDART00000157148 |
cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr2_+_21356242 | 0.22 |
ENSDART00000145670
ENSDART00000146600 |
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr3_-_25107802 | 0.22 |
ENSDART00000055432
|
rbx1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr16_+_54829293 | 0.19 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr1_+_26105141 | 0.18 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr11_-_7156620 | 0.18 |
ENSDART00000172823
ENSDART00000172879 ENSDART00000078916 |
smim7
|
small integral membrane protein 7 |
| chr6_-_41079209 | 0.17 |
ENSDART00000151592
|
rab44
|
RAB44, member RAS oncogene family |
| chr25_+_34915576 | 0.16 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr7_+_22981909 | 0.16 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr24_+_15655069 | 0.16 |
ENSDART00000042943
ENSDART00000181296 |
fbxo15
|
F-box protein 15 |
| chr3_-_43954343 | 0.16 |
ENSDART00000157580
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr6_+_13201358 | 0.14 |
ENSDART00000190290
|
CT009620.1
|
|
| chr6_+_39290777 | 0.14 |
ENSDART00000155700
|
si:dkey-195m11.11
|
si:dkey-195m11.11 |
| chr17_-_12764360 | 0.13 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr7_+_4694924 | 0.11 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr15_+_1134870 | 0.10 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr10_-_2788668 | 0.08 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr14_+_44804326 | 0.07 |
ENSDART00000079866
ENSDART00000172974 |
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr20_-_25445237 | 0.06 |
ENSDART00000145034
ENSDART00000018049 |
mrpl19
|
mitochondrial ribosomal protein L19 |
| chr1_-_17711636 | 0.05 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr6_-_14040136 | 0.05 |
ENSDART00000065361
ENSDART00000179765 |
etv5b
|
ets variant 5b |
| chr4_-_17680493 | 0.03 |
ENSDART00000180131
|
BX890572.2
|
|
| chr13_-_18069421 | 0.02 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr9_-_3519253 | 0.02 |
ENSDART00000169586
|
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr7_+_4938463 | 0.02 |
ENSDART00000143722
|
si:dkey-28d5.14
|
si:dkey-28d5.14 |
| chr22_-_36750589 | 0.01 |
ENSDART00000010824
|
acy1
|
aminoacylase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd10a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.3 | 1.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.3 | 1.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.3 | 0.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.0 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.2 | 0.6 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.2 | 1.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.4 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.1 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 1.9 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.6 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.1 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.0 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 1.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.8 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 1.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 2.0 | GO:0009749 | response to glucose(GO:0009749) |
| 0.1 | 2.8 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 1.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 5.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0090660 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 1.2 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.8 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.5 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0051262 | protein tetramerization(GO:0051262) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.6 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.2 | 1.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 1.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 0.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.2 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 1.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.3 | 1.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 0.8 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.2 | 1.0 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.2 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 1.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.7 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 3.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.0 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0016248 | calcium channel regulator activity(GO:0005246) ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 2.5 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.6 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 2.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |