Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
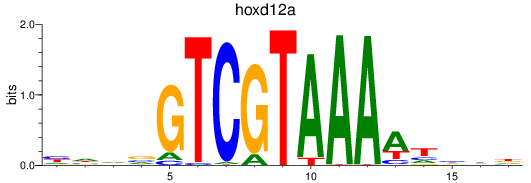
Results for hoxd12a
Z-value: 0.86
Transcription factors associated with hoxd12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd12a
|
ENSDARG00000059263 | homeobox D12a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd12a | dr11_v1_chr9_-_1986014_1986014 | -0.18 | 7.9e-02 | Click! |
Activity profile of hoxd12a motif
Sorted Z-values of hoxd12a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_30485009 | 8.98 |
ENSDART00000025698
|
zgc:153311
|
zgc:153311 |
| chr17_+_42274825 | 7.74 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr1_+_45056371 | 7.71 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr22_+_22010128 | 6.34 |
ENSDART00000172610
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr24_-_17029374 | 6.21 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr7_+_49681040 | 6.10 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr19_+_19989380 | 6.05 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr16_-_9675982 | 6.05 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr13_+_28747934 | 5.81 |
ENSDART00000160176
ENSDART00000101633 ENSDART00000136318 |
prom2
|
prominin 2 |
| chr13_-_34862452 | 5.78 |
ENSDART00000134573
ENSDART00000047552 |
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr3_-_50147160 | 5.60 |
ENSDART00000191341
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr8_-_36399884 | 5.48 |
ENSDART00000108538
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr17_-_8886735 | 4.61 |
ENSDART00000121997
|
nkl.3
|
NK-lysin tandem duplicate 3 |
| chr2_-_43635777 | 4.52 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr16_-_31754102 | 4.38 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr2_-_36914281 | 4.28 |
ENSDART00000008322
|
si:dkey-193b15.5
|
si:dkey-193b15.5 |
| chr5_-_23696926 | 3.97 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr7_-_25133783 | 3.90 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr24_-_26632171 | 3.54 |
ENSDART00000008374
ENSDART00000017384 |
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr11_+_24716837 | 3.26 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr5_-_23783739 | 3.17 |
ENSDART00000139502
|
GBGT1 (1 of many)
|
si:ch211-287c22.1 |
| chr3_-_50146854 | 3.16 |
ENSDART00000157047
|
si:dkey-120c6.5
|
si:dkey-120c6.5 |
| chr10_-_20357013 | 3.01 |
ENSDART00000080143
|
sfrp1b
|
secreted frizzled-related protein 1b |
| chr5_+_23598364 | 3.00 |
ENSDART00000132155
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr18_+_8812549 | 2.97 |
ENSDART00000017619
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr8_-_12432604 | 2.96 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr22_+_10440991 | 2.86 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr1_+_58282449 | 2.69 |
ENSDART00000131475
|
si:dkey-222h21.7
|
si:dkey-222h21.7 |
| chr16_-_47426482 | 2.63 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr8_+_23861461 | 2.60 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr1_+_36612660 | 2.58 |
ENSDART00000190784
|
ednraa
|
endothelin receptor type Aa |
| chr3_+_34140507 | 2.55 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr19_-_1948236 | 2.54 |
ENSDART00000163344
|
znrf2a
|
zinc and ring finger 2a |
| chr21_-_40174647 | 2.54 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr3_+_23063718 | 2.53 |
ENSDART00000140225
ENSDART00000184431 |
b4galnt2.1
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 1 |
| chr25_-_21066136 | 2.52 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr5_+_23599259 | 2.43 |
ENSDART00000138902
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr20_+_34845672 | 2.43 |
ENSDART00000128895
|
emilin1a
|
elastin microfibril interfacer 1a |
| chr24_-_26715174 | 2.43 |
ENSDART00000079726
|
pld1b
|
phospholipase D1b |
| chr24_-_33284945 | 2.42 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr19_+_28291062 | 2.41 |
ENSDART00000163382
|
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr15_-_28904371 | 2.40 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr2_+_31330358 | 2.36 |
ENSDART00000178066
|
clul1
|
clusterin-like 1 (retinal) |
| chr13_-_9875538 | 2.33 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr9_+_20483846 | 2.29 |
ENSDART00000192067
ENSDART00000145111 |
parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr16_+_32014552 | 2.25 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr1_+_31019165 | 2.22 |
ENSDART00000102210
|
itga6b
|
integrin, alpha 6b |
| chr8_+_20994433 | 2.22 |
ENSDART00000141736
|
si:dkeyp-82a1.8
|
si:dkeyp-82a1.8 |
| chr1_-_38171648 | 2.20 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr9_-_9242777 | 2.20 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr1_+_49568335 | 2.18 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr3_+_16663373 | 2.15 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr6_+_42338309 | 2.08 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr19_+_7424347 | 2.06 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr1_-_26782573 | 2.05 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr5_-_26764880 | 2.04 |
ENSDART00000140392
ENSDART00000134728 |
rnf181
|
ring finger protein 181 |
| chr11_-_18557929 | 2.00 |
ENSDART00000110882
ENSDART00000181381 ENSDART00000189312 |
dido1
|
death inducer-obliterator 1 |
| chr16_-_10316359 | 1.99 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr14_+_989733 | 1.96 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr6_-_13376060 | 1.95 |
ENSDART00000104732
ENSDART00000125271 |
arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6 |
| chr6_-_34838397 | 1.93 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr20_-_26929685 | 1.92 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr17_-_31695217 | 1.92 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr14_-_10387377 | 1.90 |
ENSDART00000145118
|
commd5
|
COMM domain containing 5 |
| chr11_+_20896122 | 1.90 |
ENSDART00000162339
ENSDART00000181111 |
CABZ01008730.1
|
|
| chr7_+_7570701 | 1.87 |
ENSDART00000172774
|
si:ch211-42c5.5
|
si:ch211-42c5.5 |
| chr25_+_32473277 | 1.87 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr6_-_2154137 | 1.86 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr10_-_31805923 | 1.84 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr15_+_8767650 | 1.84 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr2_-_37458527 | 1.83 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr22_-_10502780 | 1.81 |
ENSDART00000136961
|
ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr2_+_4402765 | 1.79 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr25_+_3294150 | 1.79 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr7_-_42206720 | 1.78 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr9_-_2892250 | 1.76 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr16_+_33143503 | 1.76 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr6_-_47953829 | 1.75 |
ENSDART00000157264
|
si:dkey-166d12.2
|
si:dkey-166d12.2 |
| chr21_-_22543611 | 1.74 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr21_+_45238533 | 1.73 |
ENSDART00000151481
|
si:ch73-269m14.4
|
si:ch73-269m14.4 |
| chr16_+_42471455 | 1.72 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr12_-_29233738 | 1.71 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr10_+_26612321 | 1.70 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr19_+_28291376 | 1.70 |
ENSDART00000139433
ENSDART00000103855 |
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr24_-_24796583 | 1.69 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr15_+_29116063 | 1.68 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr2_-_10338759 | 1.68 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr16_+_33144306 | 1.66 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr25_+_18964782 | 1.66 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr7_+_55292959 | 1.63 |
ENSDART00000147539
ENSDART00000073555 |
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr6_+_18321986 | 1.63 |
ENSDART00000190871
|
card14
|
caspase recruitment domain family, member 14 |
| chr8_-_31075015 | 1.60 |
ENSDART00000010993
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr22_-_15671525 | 1.56 |
ENSDART00000080286
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr19_+_29808699 | 1.55 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr18_-_41161828 | 1.53 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr18_-_2433011 | 1.53 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr4_-_25271455 | 1.50 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr25_-_37084032 | 1.49 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr19_-_31007417 | 1.49 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr9_+_41080029 | 1.45 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr16_+_9713850 | 1.44 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr22_-_10440688 | 1.43 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr12_-_17199381 | 1.42 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr5_+_32490595 | 1.41 |
ENSDART00000165417
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr22_-_31517300 | 1.39 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr17_+_30843881 | 1.37 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr14_+_16287968 | 1.36 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr11_+_2554856 | 1.35 |
ENSDART00000173000
|
zgc:113184
|
zgc:113184 |
| chr22_-_7539414 | 1.32 |
ENSDART00000159083
|
BX511034.5
|
|
| chr21_-_3007412 | 1.31 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr19_+_7595924 | 1.30 |
ENSDART00000140083
|
s100s
|
S100 calcium binding protein S |
| chr2_-_55797318 | 1.26 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr22_-_9736050 | 1.25 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr12_+_46696867 | 1.25 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
| chr24_-_5713799 | 1.24 |
ENSDART00000137293
|
dia1b
|
deleted in autism 1b |
| chr16_+_33144112 | 1.22 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr19_+_29808471 | 1.21 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr15_-_28262632 | 1.20 |
ENSDART00000134601
ENSDART00000175022 |
prpf8
|
pre-mRNA processing factor 8 |
| chr13_-_36050303 | 1.17 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr2_-_37874647 | 1.16 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr6_-_14038804 | 1.16 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr11_-_16115804 | 1.15 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr8_-_25327809 | 1.14 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr11_-_6880725 | 1.13 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr7_-_41014773 | 1.11 |
ENSDART00000013785
|
insig1
|
insulin induced gene 1 |
| chr16_-_43065164 | 1.11 |
ENSDART00000149431
|
sema4aa
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Aa |
| chr19_+_30884960 | 1.11 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr7_-_18508815 | 1.10 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr5_-_31856681 | 1.10 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr17_-_23727978 | 1.10 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr3_-_27868183 | 1.09 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr25_-_35996141 | 1.09 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr17_+_17764979 | 1.09 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr13_+_39182099 | 1.08 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr6_+_33885828 | 1.06 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr20_-_20355577 | 1.06 |
ENSDART00000018500
|
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr11_+_25560632 | 1.05 |
ENSDART00000033914
|
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr6_+_18321627 | 1.05 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr1_-_56213723 | 1.05 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr16_-_10071886 | 1.05 |
ENSDART00000136168
ENSDART00000130382 |
ptpn2a
|
protein tyrosine phosphatase, non-receptor type 2, a |
| chr19_-_35400819 | 1.04 |
ENSDART00000148080
|
rnf19b
|
ring finger protein 19B |
| chr7_+_9308625 | 1.03 |
ENSDART00000084598
|
selenos
|
selenoprotein S |
| chr6_-_57476465 | 1.02 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr2_-_22966076 | 1.02 |
ENSDART00000143412
ENSDART00000146014 ENSDART00000183443 ENSDART00000191056 ENSDART00000183539 |
sap130b
|
Sin3A-associated protein b |
| chr7_-_30624435 | 1.01 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr3_-_27065477 | 1.00 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr16_+_4078608 | 1.00 |
ENSDART00000166241
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr5_-_69499486 | 0.99 |
ENSDART00000023983
ENSDART00000180293 |
psat1
|
phosphoserine aminotransferase 1 |
| chr7_-_55051692 | 0.99 |
ENSDART00000170637
|
tpcn2
|
two pore segment channel 2 |
| chr21_+_26720803 | 0.98 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr6_-_18228358 | 0.98 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr5_+_38744660 | 0.97 |
ENSDART00000180506
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr22_+_20169352 | 0.96 |
ENSDART00000169055
ENSDART00000061617 |
hmg20b
|
high mobility group 20B |
| chr21_+_11865972 | 0.96 |
ENSDART00000081676
|
ubap1
|
ubiquitin associated protein 1 |
| chr9_-_2892045 | 0.95 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr23_+_39611688 | 0.95 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr16_+_42481447 | 0.95 |
ENSDART00000037401
|
herpud2
|
HERPUD family member 2 |
| chr16_-_33650578 | 0.92 |
ENSDART00000058460
|
utp11l
|
UTP11-like, U3 small nucleolar ribonucleoprotein (yeast) |
| chr1_-_16507812 | 0.91 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr12_+_2677303 | 0.89 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr9_+_500052 | 0.86 |
ENSDART00000166707
|
CU984600.1
|
|
| chr20_+_38837238 | 0.85 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr7_-_26125092 | 0.83 |
ENSDART00000079364
|
snapc2
|
small nuclear RNA activating complex, polypeptide 2 |
| chr7_-_2116512 | 0.83 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr10_-_3295197 | 0.82 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr18_+_3140682 | 0.82 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr11_-_36051004 | 0.81 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr16_-_31452416 | 0.80 |
ENSDART00000140880
ENSDART00000008297 ENSDART00000147373 |
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr14_+_8940326 | 0.78 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr17_+_6217704 | 0.78 |
ENSDART00000129100
|
BX000363.1
|
|
| chr6_-_53334259 | 0.78 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr7_+_20524064 | 0.78 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr23_+_33752275 | 0.77 |
ENSDART00000007260
|
si:ch211-210c8.6
|
si:ch211-210c8.6 |
| chr25_+_3788443 | 0.76 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr17_+_49500820 | 0.76 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr4_-_39448829 | 0.76 |
ENSDART00000168526
|
si:dkey-261o4.9
|
si:dkey-261o4.9 |
| chr25_-_35169303 | 0.75 |
ENSDART00000193240
|
ano9a
|
anoctamin 9a |
| chr12_-_10038870 | 0.75 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr18_+_41542542 | 0.74 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr15_-_44275069 | 0.74 |
ENSDART00000124980
|
CU929391.2
|
|
| chr19_-_7272921 | 0.73 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr5_-_43935119 | 0.73 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr4_+_76441200 | 0.73 |
ENSDART00000125751
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr10_+_25947946 | 0.72 |
ENSDART00000064393
|
ufm1
|
ubiquitin-fold modifier 1 |
| chr5_+_32490238 | 0.72 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr11_+_25560072 | 0.71 |
ENSDART00000124131
ENSDART00000147179 |
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr18_+_13275735 | 0.70 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr8_+_52442785 | 0.70 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr11_+_2416064 | 0.70 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr6_+_49742164 | 0.70 |
ENSDART00000024578
|
npepl1
|
aminopeptidase like 1 |
| chr18_-_17087138 | 0.69 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr25_-_20024829 | 0.69 |
ENSDART00000140182
ENSDART00000174776 |
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr19_-_7321221 | 0.68 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr2_-_13254821 | 0.67 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr17_+_38295847 | 0.66 |
ENSDART00000008532
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr3_-_34599662 | 0.65 |
ENSDART00000055259
|
nmrk1
|
nicotinamide riboside kinase 1 |
| chr5_+_27434601 | 0.62 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr3_+_36284986 | 0.60 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr13_+_37022601 | 0.60 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr1_+_45671687 | 0.59 |
ENSDART00000146101
|
mcoln1a
|
mucolipin 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd12a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.7 | 2.0 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.6 | 2.5 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 1.9 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.6 | 2.4 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.6 | 2.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.5 | 5.8 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.5 | 1.5 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.5 | 2.9 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.5 | 2.9 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 1.7 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 2.6 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.4 | 3.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.4 | 2.8 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.4 | 1.1 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.4 | 3.9 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.3 | 1.4 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.3 | 1.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 1.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.3 | 1.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.3 | 1.6 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 0.7 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 1.8 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.2 | 0.9 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 1.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.2 | 1.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.2 | 1.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 0.8 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.2 | 1.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 2.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.5 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.2 | 1.8 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 0.8 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 0.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 4.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.7 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 2.1 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.1 | 3.0 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.7 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 2.4 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 1.0 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.1 | 2.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 10.2 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.1 | 0.8 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.4 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 2.2 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.7 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 0.7 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 1.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 1.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.5 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 2.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 2.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.6 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.7 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 1.0 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 1.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 2.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 3.0 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.7 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.7 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 6.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.4 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 4.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 2.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.7 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 4.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 6.9 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 2.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.0 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 4.0 | GO:0006511 | ubiquitin-dependent protein catabolic process(GO:0006511) |
| 0.0 | 2.2 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.4 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 1.0 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 1.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.7 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 2.0 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 0.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 8.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 4.4 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.0 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 1.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.8 | GO:0071914 | prominosome(GO:0071914) |
| 0.6 | 5.8 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.6 | 2.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.5 | 2.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.4 | 6.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 1.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 5.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.3 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 4.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 2.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 1.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 1.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 5.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 3.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.7 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 3.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 4.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 6.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 4.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.7 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 2.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.8 | 5.8 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.8 | 4.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.8 | 6.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.6 | 1.9 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.5 | 2.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.5 | 1.5 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.5 | 3.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 1.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 6.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 1.2 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.4 | 2.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 2.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 1.0 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 1.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.3 | 1.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.3 | 2.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 6.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 2.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 2.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 4.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 2.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.7 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 5.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 0.5 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 2.2 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.7 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 1.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.3 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.7 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 3.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.6 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 3.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 1.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 4.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.0 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 2.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.5 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.1 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 1.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 1.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 13.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 2.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 5.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 3.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.8 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleoside transmembrane transporter activity(GO:0015211) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 1.4 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 1.4 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 5.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 3.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.7 | 4.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.6 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 2.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 6.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 1.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 3.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.8 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |