Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
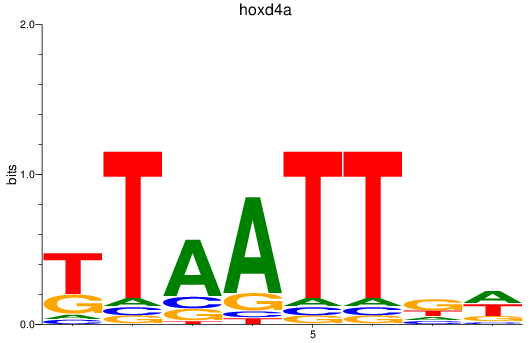
Results for hoxd4a
Z-value: 0.43
Transcription factors associated with hoxd4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd4a
|
ENSDARG00000059276 | homeobox D4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd4a | dr11_v1_chr9_-_1951144_1951144 | -0.02 | 8.7e-01 | Click! |
Activity profile of hoxd4a motif
Sorted Z-values of hoxd4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_2495197 | 3.33 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr24_+_38301080 | 2.97 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr1_-_18811517 | 2.43 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr17_-_2578026 | 2.26 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr25_+_31277415 | 2.25 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr7_-_38698583 | 1.92 |
ENSDART00000173900
ENSDART00000126737 |
cd59
|
CD59 molecule (CD59 blood group) |
| chr25_+_31276842 | 1.84 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr19_-_3167729 | 1.74 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr24_-_9985019 | 1.70 |
ENSDART00000193536
ENSDART00000189595 |
zgc:171977
|
zgc:171977 |
| chr7_-_20582842 | 1.54 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr11_+_18183220 | 1.54 |
ENSDART00000113468
|
LO018315.10
|
|
| chr24_-_9979342 | 1.52 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr3_+_24361096 | 1.39 |
ENSDART00000132387
|
pvalb6
|
parvalbumin 6 |
| chr4_+_7841627 | 1.39 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr7_-_24699985 | 1.17 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr4_-_14915268 | 1.09 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr6_-_46861676 | 1.07 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr4_-_4387012 | 1.06 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr10_-_7756865 | 1.05 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr13_-_9318891 | 1.04 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr20_-_29474859 | 0.93 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr22_+_25774750 | 0.92 |
ENSDART00000174421
|
AL929192.1
|
|
| chr10_-_26202766 | 0.89 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr23_+_19590006 | 0.86 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr4_+_14981854 | 0.81 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr2_+_56657804 | 0.78 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr18_+_29156827 | 0.75 |
ENSDART00000137587
ENSDART00000135633 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr22_-_10470663 | 0.74 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr14_-_33894915 | 0.71 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr17_+_23298928 | 0.70 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr19_-_47527093 | 0.70 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr2_+_20605186 | 0.65 |
ENSDART00000128505
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr7_+_44802353 | 0.63 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr14_-_35462148 | 0.62 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr3_+_26123736 | 0.62 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr19_+_24872159 | 0.61 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr13_-_7567707 | 0.60 |
ENSDART00000190296
ENSDART00000180348 |
pitx3
|
paired-like homeodomain 3 |
| chr23_-_21453614 | 0.58 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr17_-_4252221 | 0.57 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr6_+_21001264 | 0.55 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr3_-_32337653 | 0.55 |
ENSDART00000156918
ENSDART00000156551 |
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr16_-_55028740 | 0.51 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr8_+_3820134 | 0.51 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr24_+_15020402 | 0.51 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr16_+_13818500 | 0.50 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr23_-_31913069 | 0.50 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr11_+_25259058 | 0.50 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr9_+_29548195 | 0.49 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr15_+_23657051 | 0.48 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr6_+_36839509 | 0.47 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr22_+_38037530 | 0.47 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr14_+_26247319 | 0.46 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr25_+_3318192 | 0.45 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr23_-_31913231 | 0.45 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr3_-_49382896 | 0.45 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr12_-_34435604 | 0.44 |
ENSDART00000115088
|
birc5a
|
baculoviral IAP repeat containing 5a |
| chr7_+_20017211 | 0.44 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr15_-_30816370 | 0.44 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr20_+_23173710 | 0.41 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr8_+_17069577 | 0.40 |
ENSDART00000131847
|
si:dkey-190g11.3
|
si:dkey-190g11.3 |
| chr14_-_30905288 | 0.40 |
ENSDART00000173449
ENSDART00000173451 |
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr22_+_8753092 | 0.40 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr23_-_26652273 | 0.39 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr4_-_14315855 | 0.39 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr3_-_31258459 | 0.39 |
ENSDART00000177928
|
LO017965.1
|
|
| chr22_+_8753365 | 0.39 |
ENSDART00000106086
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr8_-_34065573 | 0.39 |
ENSDART00000186946
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr6_-_44402358 | 0.39 |
ENSDART00000193007
ENSDART00000193603 |
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr10_+_21522545 | 0.38 |
ENSDART00000131097
|
BX957322.1
|
|
| chr2_-_45135591 | 0.38 |
ENSDART00000014691
ENSDART00000123916 |
eif4e2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr3_+_27770110 | 0.37 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr10_+_39952995 | 0.37 |
ENSDART00000183077
|
BX927333.1
|
|
| chr25_-_10503043 | 0.37 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr25_-_13839743 | 0.37 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr5_-_30491172 | 0.36 |
ENSDART00000138464
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr9_+_31534601 | 0.35 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr2_+_45362016 | 0.34 |
ENSDART00000139154
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr8_+_28467893 | 0.34 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr9_-_22918413 | 0.34 |
ENSDART00000007392
|
arl5a
|
ADP-ribosylation factor-like 5A |
| chr16_-_41465542 | 0.33 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr2_+_4146299 | 0.32 |
ENSDART00000173418
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr20_+_18551657 | 0.32 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr25_+_34888886 | 0.32 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr4_+_15944245 | 0.32 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr16_+_29509133 | 0.31 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr14_+_15620640 | 0.31 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr24_-_21819010 | 0.31 |
ENSDART00000091096
|
CR352265.1
|
|
| chr23_+_404975 | 0.30 |
ENSDART00000181336
|
CU929146.1
|
|
| chr16_-_15988320 | 0.30 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr2_+_16696052 | 0.30 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr2_+_33326522 | 0.30 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr22_+_28818291 | 0.30 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr5_+_36611128 | 0.30 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr7_-_5896339 | 0.30 |
ENSDART00000172908
|
si:dkey-23a13.2
|
si:dkey-23a13.2 |
| chr8_+_52284075 | 0.29 |
ENSDART00000098439
|
ube2d4
|
ubiquitin-conjugating enzyme E2D 4 (putative) |
| chr4_-_18202291 | 0.28 |
ENSDART00000169704
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr18_-_16392229 | 0.28 |
ENSDART00000189031
|
mgat4c
|
mgat4 family, member C |
| chr18_-_48296793 | 0.28 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr21_+_9689103 | 0.28 |
ENSDART00000165132
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr23_-_24343363 | 0.27 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr23_-_36823932 | 0.27 |
ENSDART00000142305
|
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr22_+_37631234 | 0.27 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr20_+_30939178 | 0.27 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr15_-_34658057 | 0.26 |
ENSDART00000110964
|
bag6
|
BCL2 associated athanogene 6 |
| chr8_-_50888806 | 0.26 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr15_-_45110011 | 0.25 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr7_-_73851280 | 0.25 |
ENSDART00000190053
|
FP236812.3
|
|
| chr2_+_37815687 | 0.25 |
ENSDART00000166352
|
sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr16_-_17541890 | 0.24 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr22_-_2922053 | 0.24 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr18_+_33310811 | 0.23 |
ENSDART00000131587
|
v2ra16
|
vomeronasal 2 receptor, a16 |
| chr11_-_30611814 | 0.23 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr18_+_34225520 | 0.22 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr11_-_27874116 | 0.22 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr6_-_15641686 | 0.22 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr16_+_23403602 | 0.22 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr11_-_40101246 | 0.22 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr21_+_37445871 | 0.22 |
ENSDART00000076333
|
pgk1
|
phosphoglycerate kinase 1 |
| chr11_-_22303678 | 0.22 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr16_+_14707960 | 0.22 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr9_+_34380299 | 0.21 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr24_+_26134209 | 0.21 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr6_+_45692026 | 0.21 |
ENSDART00000164759
|
cntn4
|
contactin 4 |
| chr1_+_18811679 | 0.21 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr4_+_65127317 | 0.21 |
ENSDART00000166475
|
znf1126
|
zinc finger protein 1126 |
| chr22_-_12337781 | 0.21 |
ENSDART00000188357
ENSDART00000123574 |
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr19_-_47455944 | 0.21 |
ENSDART00000190005
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr13_+_1726953 | 0.21 |
ENSDART00000103004
|
zmp:0000000760
|
zmp:0000000760 |
| chr7_-_26532089 | 0.21 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr5_+_66433287 | 0.21 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr25_-_2723682 | 0.20 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr18_+_16192083 | 0.20 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr13_+_11440389 | 0.20 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr11_-_28911172 | 0.19 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr22_-_19552796 | 0.19 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr5_+_24520437 | 0.19 |
ENSDART00000144560
|
dgcr8
|
DGCR8 microprocessor complex subunit |
| chr5_-_68795063 | 0.19 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr1_+_50921266 | 0.19 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr1_-_44157273 | 0.19 |
ENSDART00000166618
|
slc43a3a
|
solute carrier family 43, member 3a |
| chr24_+_37902687 | 0.18 |
ENSDART00000042273
|
ccdc78
|
coiled-coil domain containing 78 |
| chr8_-_44004135 | 0.18 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr24_-_1021318 | 0.18 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr2_-_23677422 | 0.18 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr12_+_47907277 | 0.18 |
ENSDART00000097714
|
tbata
|
thymus, brain and testes associated |
| chr7_-_20758825 | 0.18 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr1_+_23784905 | 0.17 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr1_-_45213565 | 0.17 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr24_-_13364311 | 0.17 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr14_-_32089117 | 0.17 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr17_+_32343121 | 0.17 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr18_-_5392667 | 0.17 |
ENSDART00000169896
|
CABZ01084603.1
|
|
| chr24_-_35707552 | 0.16 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_31618243 | 0.16 |
ENSDART00000016592
|
mmgt1
|
membrane magnesium transporter 1 |
| chr3_+_32691275 | 0.16 |
ENSDART00000191838
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr24_-_3426620 | 0.16 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr5_-_31856681 | 0.15 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr25_+_30074947 | 0.15 |
ENSDART00000154849
|
si:ch211-147k10.5
|
si:ch211-147k10.5 |
| chr8_+_8238727 | 0.15 |
ENSDART00000144838
|
plxnb3
|
plexin B3 |
| chr23_+_4689626 | 0.15 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr11_-_36341189 | 0.15 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr22_+_37631034 | 0.15 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr23_-_16980213 | 0.14 |
ENSDART00000046889
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr23_-_44207349 | 0.14 |
ENSDART00000186276
|
zgc:158659
|
zgc:158659 |
| chr25_-_1348370 | 0.14 |
ENSDART00000154865
|
itga11b
|
integrin, alpha 11b |
| chr8_+_28257219 | 0.14 |
ENSDART00000183749
|
BX664742.3
|
|
| chr7_+_5910467 | 0.14 |
ENSDART00000173232
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr3_+_19460991 | 0.14 |
ENSDART00000169124
|
pde6ga
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog a |
| chr11_-_35171768 | 0.13 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr17_-_15201031 | 0.13 |
ENSDART00000124426
|
socs4
|
suppressor of cytokine signaling 4 |
| chr6_-_39270851 | 0.13 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr13_+_48359573 | 0.13 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr19_+_4139065 | 0.13 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr3_+_21225750 | 0.13 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr6_+_21992820 | 0.13 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr5_+_42400777 | 0.13 |
ENSDART00000183114
|
BX548073.8
|
|
| chr2_+_4146606 | 0.13 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr1_+_56919742 | 0.13 |
ENSDART00000152241
|
si:ch211-1f22.9
|
si:ch211-1f22.9 |
| chr6_-_14292307 | 0.13 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr6_+_30689239 | 0.12 |
ENSDART00000065206
|
wdr78
|
WD repeat domain 78 |
| chr2_+_2181709 | 0.12 |
ENSDART00000092763
ENSDART00000148553 |
ccdc13
|
coiled-coil domain containing 13 |
| chr21_+_4256291 | 0.12 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr10_-_40484531 | 0.12 |
ENSDART00000142464
ENSDART00000180318 |
taar20z
|
trace amine associated receptor 20z |
| chr16_+_8695595 | 0.12 |
ENSDART00000173249
|
si:cabz01093077.1
|
si:cabz01093077.1 |
| chr19_+_32979331 | 0.12 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr12_+_18681477 | 0.11 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr13_+_40034176 | 0.11 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr18_+_30508729 | 0.11 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr4_+_9028819 | 0.11 |
ENSDART00000102893
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr10_-_24759616 | 0.11 |
ENSDART00000079528
|
ilk
|
integrin-linked kinase |
| chr2_+_9755422 | 0.11 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr23_-_14990865 | 0.11 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr9_+_48219111 | 0.11 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr4_-_65525733 | 0.11 |
ENSDART00000159401
|
BX293993.1
|
|
| chr2_+_31614533 | 0.11 |
ENSDART00000111038
ENSDART00000181311 |
si:ch211-106h4.6
|
si:ch211-106h4.6 |
| chr19_-_1855368 | 0.11 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr20_-_16171297 | 0.10 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr3_+_25825043 | 0.10 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr9_-_52490579 | 0.10 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr2_+_33726862 | 0.10 |
ENSDART00000146745
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr1_+_8442071 | 0.10 |
ENSDART00000143547
|
myo15ab
|
myosin XVAb |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.2 | 1.7 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.2 | 3.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 0.5 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 2.3 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.6 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 1.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 4.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 1.0 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.4 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 1.6 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.4 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.6 | GO:0021984 | adenohypophysis development(GO:0021984) lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.4 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 3.3 | GO:0060537 | muscle tissue development(GO:0060537) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 4.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.2 | 2.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.8 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 3.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0043531 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |