Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
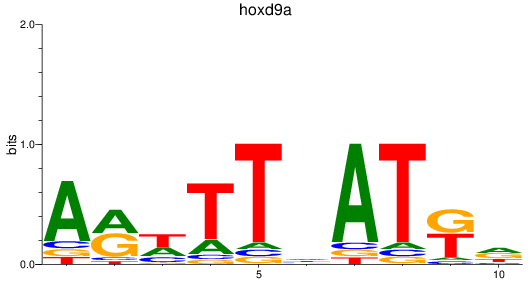
Results for hoxd9a
Z-value: 0.91
Transcription factors associated with hoxd9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd9a
|
ENSDARG00000059274 | homeobox D9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd9a | dr11_v1_chr9_-_1965727_1965727 | -0.05 | 6.6e-01 | Click! |
Activity profile of hoxd9a motif
Sorted Z-values of hoxd9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_46425262 | 15.31 |
ENSDART00000153690
|
sc:d0202
|
sc:d0202 |
| chr6_-_53426773 | 13.92 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr13_+_17672527 | 13.51 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr16_+_23913943 | 12.43 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr11_+_37201483 | 12.08 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr5_+_28830643 | 11.86 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr15_+_14856307 | 11.07 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr22_+_37888249 | 9.84 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr22_-_24880824 | 9.40 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr14_+_32430982 | 8.43 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr22_+_12770877 | 7.76 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr23_+_37482727 | 7.73 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr14_+_29975268 | 7.25 |
ENSDART00000172985
|
cyp4v7
|
cytochrome P450, family 4, subfamily V, polypeptide 7 |
| chr24_-_38083378 | 6.90 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr7_-_26308098 | 6.58 |
ENSDART00000146440
ENSDART00000146935 ENSDART00000164627 |
zgc:77439
|
zgc:77439 |
| chr7_-_26306546 | 6.55 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr3_+_1030479 | 6.43 |
ENSDART00000143449
|
zgc:172253
|
zgc:172253 |
| chr7_+_56577522 | 6.01 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr21_-_42831033 | 5.62 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr17_+_20174812 | 5.26 |
ENSDART00000186628
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr15_-_32383340 | 5.23 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr4_+_14900042 | 5.04 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr6_-_58764672 | 4.91 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr10_+_38708099 | 4.75 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr2_+_11031360 | 4.65 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr2_-_11119303 | 4.54 |
ENSDART00000135450
ENSDART00000131836 |
cryz
|
crystallin, zeta (quinone reductase) |
| chr25_-_30383617 | 4.53 |
ENSDART00000165342
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr25_+_23280220 | 4.43 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr21_+_39963851 | 4.30 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr6_+_8626427 | 4.17 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr16_+_1383914 | 4.10 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr3_+_21225750 | 4.08 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr6_-_7776612 | 3.99 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr5_+_57480014 | 3.93 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr5_-_1963498 | 3.91 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr19_+_2876106 | 3.82 |
ENSDART00000189309
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr4_+_14899720 | 3.81 |
ENSDART00000187456
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr20_-_33515890 | 3.70 |
ENSDART00000159987
|
si:dkey-65b13.13
|
si:dkey-65b13.13 |
| chr12_-_990149 | 3.70 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr22_-_17671348 | 3.70 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr11_+_43431289 | 3.61 |
ENSDART00000192526
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr21_+_39941875 | 3.56 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr2_+_27855346 | 3.49 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr6_+_41191482 | 3.45 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr6_-_52675630 | 3.44 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr23_+_7379728 | 3.43 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr15_-_32383529 | 3.36 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr23_+_2666944 | 3.33 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr22_-_16154771 | 3.22 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr1_+_38858399 | 3.12 |
ENSDART00000165454
|
CU915762.1
|
|
| chr19_+_10855158 | 3.07 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr3_+_60277300 | 3.01 |
ENSDART00000170977
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr22_+_1028724 | 2.95 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr16_-_28658341 | 2.95 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr19_-_657439 | 2.91 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr7_-_16194952 | 2.84 |
ENSDART00000173739
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr7_+_8358547 | 2.83 |
ENSDART00000173016
|
jac7
|
jacalin 7 |
| chr17_-_20236228 | 2.76 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr10_-_44467384 | 2.60 |
ENSDART00000186382
|
CABZ01072089.1
|
|
| chr8_+_22359881 | 2.59 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr11_-_3366782 | 2.57 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr7_-_24046999 | 2.53 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr20_-_43741159 | 2.53 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr22_+_16555939 | 2.52 |
ENSDART00000012604
|
pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr8_-_36287046 | 2.50 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr8_+_471342 | 2.49 |
ENSDART00000167205
|
nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr13_+_29238850 | 2.47 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr12_+_1286642 | 2.45 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr7_-_51953807 | 2.41 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr2_-_39558643 | 2.36 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr20_-_52902693 | 2.35 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr3_+_46763745 | 2.35 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr7_+_66822229 | 2.35 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr13_+_45709380 | 2.34 |
ENSDART00000192862
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr24_-_1985007 | 2.34 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr2_+_11685742 | 2.34 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr11_-_26701611 | 2.31 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr12_+_3871452 | 2.29 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr15_+_15390882 | 2.27 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr22_+_2959879 | 2.24 |
ENSDART00000185354
ENSDART00000063545 |
impact
|
impact RWD domain protein |
| chr24_+_5912635 | 2.22 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr24_-_25166416 | 2.17 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr15_-_23793641 | 2.14 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr10_-_36808348 | 2.12 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr10_+_14488625 | 2.11 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr18_+_44849809 | 2.09 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr15_-_4580763 | 2.05 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr4_-_17669881 | 2.04 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr21_-_11646878 | 2.02 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr3_+_60761811 | 2.01 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr8_+_39674707 | 2.01 |
ENSDART00000126301
ENSDART00000040330 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr21_+_39941184 | 2.00 |
ENSDART00000133604
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr10_-_14488472 | 2.00 |
ENSDART00000101298
ENSDART00000138161 |
galt
|
galactose-1-phosphate uridylyltransferase |
| chr7_+_38510197 | 1.96 |
ENSDART00000173468
ENSDART00000100479 |
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr7_+_35068036 | 1.95 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr7_-_28658143 | 1.93 |
ENSDART00000173556
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr6_-_39727244 | 1.91 |
ENSDART00000065057
|
itgb7
|
integrin, beta 7 |
| chr3_+_12440099 | 1.91 |
ENSDART00000158060
|
vasnb
|
vasorin b |
| chr13_-_36535128 | 1.88 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr12_+_19199735 | 1.84 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr20_-_40760217 | 1.82 |
ENSDART00000101007
|
cx34.5
|
connexin 34.5 |
| chr13_-_24260609 | 1.82 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr7_-_24047316 | 1.81 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr20_-_21910043 | 1.80 |
ENSDART00000152290
ENSDART00000109084 |
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr2_+_11119315 | 1.80 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr23_-_26128593 | 1.76 |
ENSDART00000136855
ENSDART00000193700 ENSDART00000163984 |
rca2.2
|
regulator of complement activation group 2 gene 2 |
| chr15_+_17406920 | 1.74 |
ENSDART00000081059
|
rps6kb1b
|
ribosomal protein S6 kinase b, polypeptide 1b |
| chr12_+_1000323 | 1.74 |
ENSDART00000054363
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr2_+_27855102 | 1.73 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr24_-_21090447 | 1.71 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr6_-_2134581 | 1.70 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr21_+_39941559 | 1.69 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr22_-_9736050 | 1.69 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr4_+_8680767 | 1.68 |
ENSDART00000182726
|
adipor2
|
adiponectin receptor 2 |
| chr20_-_19858936 | 1.65 |
ENSDART00000161579
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr2_+_49713592 | 1.62 |
ENSDART00000189624
|
BX323861.3
|
|
| chr17_-_43594864 | 1.60 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr6_-_141564 | 1.59 |
ENSDART00000151245
ENSDART00000063876 |
s1pr5b
|
sphingosine-1-phosphate receptor 5b |
| chr23_-_26252103 | 1.58 |
ENSDART00000160873
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr18_-_44908479 | 1.57 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr4_-_16658514 | 1.57 |
ENSDART00000133837
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr5_-_9216758 | 1.56 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr13_-_24257631 | 1.55 |
ENSDART00000146524
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr20_+_18163821 | 1.55 |
ENSDART00000186507
|
aqp4
|
aquaporin 4 |
| chr17_+_30442518 | 1.54 |
ENSDART00000155264
|
lpin1
|
lipin 1 |
| chr6_-_34938678 | 1.53 |
ENSDART00000186689
ENSDART00000131610 |
serbp1a
|
SERPINE1 mRNA binding protein 1a |
| chr6_-_42983843 | 1.51 |
ENSDART00000130666
|
tnfrsf18
|
tumor necrosis factor receptor superfamily, member 18 |
| chr16_+_11660839 | 1.51 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr19_+_47299212 | 1.50 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr2_+_10280645 | 1.50 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr10_-_1276046 | 1.50 |
ENSDART00000169779
|
pdlim5b
|
PDZ and LIM domain 5b |
| chr8_-_39884359 | 1.50 |
ENSDART00000131372
|
mlec
|
malectin |
| chr2_-_10600950 | 1.48 |
ENSDART00000160216
|
BX323564.1
|
|
| chr23_+_4483083 | 1.46 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr20_+_53368611 | 1.45 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr16_-_16761164 | 1.45 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr9_+_21993092 | 1.44 |
ENSDART00000059693
|
crygm7
|
crystallin, gamma M7 |
| chr10_-_24784446 | 1.44 |
ENSDART00000140877
|
si:ch1073-15f19.2
|
si:ch1073-15f19.2 |
| chr10_+_14963898 | 1.41 |
ENSDART00000187363
ENSDART00000175732 |
si:dkey-88l16.3
|
si:dkey-88l16.3 |
| chr5_-_24238733 | 1.41 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr5_+_26686279 | 1.39 |
ENSDART00000193543
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr2_-_43191465 | 1.37 |
ENSDART00000025254
|
crema
|
cAMP responsive element modulator a |
| chr25_-_16768078 | 1.37 |
ENSDART00000025186
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr5_-_4216324 | 1.36 |
ENSDART00000168610
|
si:ch211-283g2.2
|
si:ch211-283g2.2 |
| chr20_+_38458084 | 1.36 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr15_-_14193926 | 1.34 |
ENSDART00000162707
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr19_+_7810028 | 1.33 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr1_+_41898919 | 1.32 |
ENSDART00000140845
|
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr24_-_3477103 | 1.32 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr25_-_23526058 | 1.30 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr19_+_4139065 | 1.30 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr11_-_19585859 | 1.29 |
ENSDART00000165989
|
atxn7
|
ataxin 7 |
| chr21_+_35327328 | 1.29 |
ENSDART00000130626
|
rars
|
arginyl-tRNA synthetase |
| chr2_+_4402765 | 1.27 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr10_-_40557210 | 1.26 |
ENSDART00000135297
|
taar18b
|
trace amine associated receptor 18b |
| chr16_+_27383717 | 1.26 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr12_+_30723778 | 1.25 |
ENSDART00000153291
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr16_-_25400257 | 1.24 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr6_+_21684296 | 1.23 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr8_-_38355268 | 1.23 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr5_+_45007962 | 1.22 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr17_-_45383699 | 1.22 |
ENSDART00000141182
|
tmem206
|
transmembrane protein 206 |
| chr21_-_32781612 | 1.22 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr15_+_11825366 | 1.21 |
ENSDART00000190042
|
prkd2
|
protein kinase D2 |
| chr15_-_23699737 | 1.20 |
ENSDART00000078311
|
zgc:154093
|
zgc:154093 |
| chr8_+_3405612 | 1.19 |
ENSDART00000163437
|
zgc:112433
|
zgc:112433 |
| chr6_+_32834007 | 1.18 |
ENSDART00000157353
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr4_-_14954327 | 1.18 |
ENSDART00000182729
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr2_+_23081402 | 1.18 |
ENSDART00000183073
|
mfsd12a
|
major facilitator superfamily domain containing 12a |
| chr22_+_35205968 | 1.17 |
ENSDART00000150467
|
tsc22d2
|
TSC22 domain family 2 |
| chr6_-_57539141 | 1.16 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr13_-_280827 | 1.15 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr16_-_7443388 | 1.15 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr4_-_14954029 | 1.14 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr8_-_36327328 | 1.14 |
ENSDART00000183333
|
zgc:103700
|
zgc:103700 |
| chr18_-_36066087 | 1.13 |
ENSDART00000059352
ENSDART00000145177 |
exosc5
|
exosome component 5 |
| chr5_+_38685089 | 1.12 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr11_-_4787083 | 1.12 |
ENSDART00000159425
|
FHIT
|
si:ch211-63i20.3 |
| chr25_-_13320986 | 1.12 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr14_+_33329420 | 1.10 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr5_-_66798729 | 1.09 |
ENSDART00000113077
|
FERMT3 (1 of many)
|
im:7154036 |
| chr10_+_23537576 | 1.09 |
ENSDART00000130515
|
CR847844.1
|
|
| chr25_+_3099073 | 1.09 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr10_-_35410518 | 1.08 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr11_-_34065718 | 1.08 |
ENSDART00000110608
|
col6a1
|
collagen, type VI, alpha 1 |
| chr2_+_19195841 | 1.08 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr17_-_14876758 | 1.08 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr1_-_17715493 | 1.08 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr2_+_42072231 | 1.07 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr16_-_46558708 | 1.07 |
ENSDART00000128037
|
si:dkey-152b24.8
|
si:dkey-152b24.8 |
| chr4_+_42175261 | 1.07 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr20_-_5229157 | 1.07 |
ENSDART00000114851
ENSDART00000182876 |
ccdc85cb
|
coiled-coil domain containing 85C, b |
| chr22_+_6084771 | 1.06 |
ENSDART00000009590
|
si:dkey-19a16.16
|
si:dkey-19a16.16 |
| chr20_+_32406011 | 1.06 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr11_+_23760470 | 1.06 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr15_+_24795473 | 1.06 |
ENSDART00000139689
ENSDART00000141033 ENSDART00000100746 ENSDART00000135677 |
gosr1
|
golgi SNAP receptor complex member 1 |
| chr17_-_27419499 | 1.05 |
ENSDART00000186773
|
ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr4_+_359970 | 1.04 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr5_-_23805387 | 1.04 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr3_-_46403778 | 1.02 |
ENSDART00000074422
|
cdip1
|
cell death-inducing p53 target 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 2.4 | 7.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 2.2 | 11.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 1.6 | 9.4 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.9 | 5.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.9 | 3.4 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.9 | 3.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.8 | 2.5 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.8 | 4.0 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.8 | 7.7 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.8 | 3.1 | GO:0097242 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.7 | 2.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.7 | 2.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.6 | 3.6 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.6 | 2.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.6 | 2.9 | GO:0006833 | water transport(GO:0006833) |
| 0.5 | 4.9 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.5 | 3.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 2.2 | GO:0010447 | response to acidic pH(GO:0010447) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.4 | 8.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.4 | 2.0 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 1.9 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.4 | 3.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.3 | 1.0 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 1.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 1.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 2.9 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.3 | 2.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.3 | 2.0 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.3 | 23.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.3 | 8.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.3 | 1.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.3 | 1.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 0.8 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.3 | 1.3 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 1.2 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.2 | 1.8 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) |
| 0.2 | 0.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 12.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.2 | 1.1 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 1.2 | GO:0089700 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.2 | 1.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.7 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 4.4 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.2 | 0.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 3.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 4.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 1.7 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 15.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 1.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 3.2 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.2 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.3 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) arginine transport(GO:0015809) |
| 0.1 | 1.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 2.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 2.9 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 2.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 2.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.1 | 0.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 11.8 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 3.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 4.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 3.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 1.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 5.7 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 16.4 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 1.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.5 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 2.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.7 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 1.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 1.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0035545 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.0 | 2.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 1.2 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 1.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 1.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 1.1 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 1.9 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 2.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 6.4 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 0.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.7 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.1 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.9 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 1.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.8 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 0.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.1 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.7 | 5.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.6 | 2.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.5 | 2.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 1.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.3 | 4.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 1.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 0.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 2.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 1.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 0.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.8 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.0 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 3.2 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 3.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 5.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 4.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 4.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 41.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 23.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 12.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 2.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 11.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.9 | 7.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.5 | 4.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 1.5 | 13.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.2 | 3.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 1.2 | 4.9 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 1.2 | 3.6 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.9 | 7.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.8 | 2.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.7 | 11.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.7 | 4.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.7 | 3.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.5 | 1.5 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.5 | 13.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.5 | 2.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.5 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 1.3 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.4 | 1.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 1.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.3 | 11.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 4.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 2.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 3.1 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.2 | 1.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.2 | 1.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 2.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 0.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 2.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 1.3 | GO:0035673 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 20.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 39.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 4.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 1.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 0.7 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 2.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.6 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 1.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.6 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.4 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 2.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 7.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 2.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 1.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 8.5 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 2.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 2.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 2.2 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.6 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 4.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 10.4 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 3.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.8 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 1.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.0 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 1.1 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 3.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 1.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 11.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 2.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 11.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 1.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 7.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 8.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 2.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 3.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 2.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 0.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 1.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.8 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.8 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 7.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 3.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 3.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 2.0 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |