Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
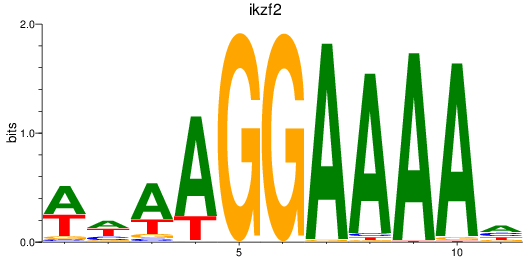
Results for ikzf2
Z-value: 1.02
Transcription factors associated with ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf2
|
ENSDARG00000069111 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | dr11_v1_chr9_-_40014339_40014339 | 0.75 | 2.2e-18 | Click! |
Activity profile of ikzf2 motif
Sorted Z-values of ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_30921246 | 19.31 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr3_+_39540014 | 13.54 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr20_+_46040666 | 13.03 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr6_-_33075576 | 10.00 |
ENSDART00000154017
|
si:dkey-170g13.2
|
si:dkey-170g13.2 |
| chr16_-_31976269 | 9.04 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr25_-_8602437 | 8.86 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr1_+_1689775 | 8.85 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 4 |
| chr18_-_17485419 | 8.28 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr12_-_8070969 | 7.89 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr18_+_7591381 | 7.66 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr15_-_46779934 | 7.49 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr21_+_11248448 | 7.24 |
ENSDART00000142431
|
arid6
|
AT-rich interaction domain 6 |
| chr16_+_23397785 | 7.19 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr10_+_36662640 | 7.09 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr15_+_46356879 | 7.02 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr16_+_23398369 | 6.87 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr15_-_33834577 | 6.36 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr12_+_17106117 | 6.28 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr9_+_45839260 | 5.82 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr12_-_15205087 | 5.74 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr18_+_19648275 | 5.72 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr13_-_21672131 | 5.68 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr1_+_9966384 | 5.47 |
ENSDART00000132607
|
si:dkeyp-75b4.8
|
si:dkeyp-75b4.8 |
| chr6_-_8489810 | 5.44 |
ENSDART00000124643
|
rasal3
|
RAS protein activator like 3 |
| chr2_+_45191049 | 5.35 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr8_-_23599096 | 5.27 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr4_+_18843015 | 5.16 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr16_+_38201840 | 5.02 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr3_+_14611299 | 4.83 |
ENSDART00000140577
|
tspan35
|
tetraspanin 35 |
| chr11_-_21404358 | 4.81 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr20_+_16881883 | 4.81 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr4_+_76659013 | 4.66 |
ENSDART00000147908
ENSDART00000134229 |
ms4a17a.5
|
membrane-spanning 4-domains, subfamily A, member 17A.5 |
| chr12_+_23424108 | 4.64 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr18_+_48428713 | 4.61 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr20_-_5291012 | 4.59 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr11_+_10548171 | 4.58 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr4_+_76775837 | 4.52 |
ENSDART00000174167
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr2_-_37465517 | 4.52 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr2_-_48196092 | 4.47 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr18_+_44703343 | 4.36 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr5_-_41049690 | 4.34 |
ENSDART00000174936
ENSDART00000135030 |
pdzd2
|
PDZ domain containing 2 |
| chr18_-_7481036 | 4.26 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr13_-_31647323 | 4.19 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr16_+_19732543 | 4.19 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr22_-_28650442 | 4.07 |
ENSDART00000019846
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr4_+_7508316 | 3.96 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr24_-_25166720 | 3.87 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr3_-_20091964 | 3.83 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr21_-_26495700 | 3.80 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr16_-_26140768 | 3.77 |
ENSDART00000143960
|
cd79a
|
CD79a molecule, immunoglobulin-associated alpha |
| chr18_+_8346920 | 3.76 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr9_+_53276356 | 3.76 |
ENSDART00000003310
|
sox21b
|
SRY (sex determining region Y)-box 21b |
| chr16_+_29492749 | 3.75 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr14_-_34605607 | 3.64 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr7_-_35432901 | 3.54 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr20_+_38276690 | 3.52 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr5_-_23574234 | 3.50 |
ENSDART00000002453
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr7_+_65673885 | 3.48 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr4_-_4261673 | 3.46 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr3_-_19495814 | 3.41 |
ENSDART00000162248
|
CU571315.3
|
|
| chr14_-_3381303 | 3.39 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr3_-_44059902 | 3.39 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr4_-_4250317 | 3.36 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr24_+_39990695 | 3.33 |
ENSDART00000040281
|
BX323854.1
|
|
| chr21_+_25187210 | 3.31 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr6_+_23809163 | 3.30 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr20_+_29436601 | 3.30 |
ENSDART00000136804
|
fmn1
|
formin 1 |
| chr12_-_4841018 | 3.30 |
ENSDART00000166500
|
zgc:163073
|
zgc:163073 |
| chr23_+_45027263 | 3.29 |
ENSDART00000058364
|
hmgb2b
|
high mobility group box 2b |
| chr16_+_13427967 | 3.27 |
ENSDART00000038196
|
zgc:101640
|
zgc:101640 |
| chr19_-_25519612 | 3.27 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr6_-_40899618 | 3.24 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr20_+_26702377 | 3.14 |
ENSDART00000077753
|
foxc1b
|
forkhead box C1b |
| chr14_-_32876280 | 3.11 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr8_+_20880848 | 3.07 |
ENSDART00000134488
ENSDART00000138605 ENSDART00000192234 |
si:ch73-196i15.3
|
si:ch73-196i15.3 |
| chr16_-_31756859 | 3.06 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr12_+_22576404 | 3.06 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr8_-_31062811 | 3.05 |
ENSDART00000142528
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr12_-_22379421 | 3.04 |
ENSDART00000187875
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr4_+_4232562 | 3.03 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr9_+_14010823 | 3.02 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr21_-_38852860 | 3.02 |
ENSDART00000166101
|
tlr22
|
toll-like receptor 22 |
| chr1_-_24462544 | 2.99 |
ENSDART00000184421
ENSDART00000184987 ENSDART00000126950 |
sh3d19
|
SH3 domain containing 19 |
| chr21_+_15346363 | 2.98 |
ENSDART00000134167
|
si:dkey-11o15.7
|
si:dkey-11o15.7 |
| chr7_+_73801377 | 2.97 |
ENSDART00000184051
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr10_-_7785930 | 2.94 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr3_+_32112004 | 2.93 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr8_+_21254192 | 2.81 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr7_+_2228276 | 2.81 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr11_-_11910225 | 2.79 |
ENSDART00000159922
|
si:ch211-69b7.6
|
si:ch211-69b7.6 |
| chr23_+_20408227 | 2.76 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr7_-_51368681 | 2.75 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr9_-_8454060 | 2.74 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr10_+_9159279 | 2.67 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr21_+_5800306 | 2.61 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr18_+_48423973 | 2.58 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr16_-_22930925 | 2.55 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr16_+_29492937 | 2.54 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr11_-_21404044 | 2.51 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr13_-_8446341 | 2.48 |
ENSDART00000080382
|
epas1b
|
endothelial PAS domain protein 1b |
| chr3_+_37707432 | 2.45 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr21_-_21781158 | 2.43 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr24_+_9744012 | 2.42 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr21_+_27189490 | 2.41 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr2_-_43151358 | 2.40 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr25_-_29074064 | 2.33 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr7_+_44593756 | 2.31 |
ENSDART00000125365
|
si:ch211-189a15.5
|
si:ch211-189a15.5 |
| chr16_-_38333976 | 2.31 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr11_-_25418856 | 2.31 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr24_+_19518570 | 2.30 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr16_-_22989262 | 2.29 |
ENSDART00000191008
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr3_+_22377312 | 2.29 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr14_-_34605804 | 2.27 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr11_+_20899029 | 2.27 |
ENSDART00000163029
|
zgc:162182
|
zgc:162182 |
| chr6_+_35362225 | 2.26 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr25_-_18470695 | 2.25 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr8_-_30242706 | 2.24 |
ENSDART00000139864
ENSDART00000143809 |
zgc:162939
|
zgc:162939 |
| chr12_+_38929663 | 2.24 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr11_+_25139495 | 2.21 |
ENSDART00000168368
|
si:ch211-25d12.7
|
si:ch211-25d12.7 |
| chr19_+_11217279 | 2.20 |
ENSDART00000181859
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr5_-_34609337 | 2.18 |
ENSDART00000145792
|
hexb
|
hexosaminidase B (beta polypeptide) |
| chr5_-_28968964 | 2.17 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr12_+_27232173 | 2.16 |
ENSDART00000193714
|
tmem106a
|
transmembrane protein 106A |
| chr20_-_26937453 | 2.16 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr9_+_33417969 | 2.15 |
ENSDART00000024795
|
gpr34b
|
G protein-coupled receptor 34b |
| chr12_+_38878830 | 2.14 |
ENSDART00000156926
|
si:ch211-39f2.3
|
si:ch211-39f2.3 |
| chr8_+_10304981 | 2.09 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr7_-_5125799 | 2.08 |
ENSDART00000173390
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr17_+_38255105 | 2.07 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr16_-_9980402 | 2.07 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr23_+_40133136 | 2.05 |
ENSDART00000157616
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr10_+_44692272 | 2.05 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr19_+_823945 | 2.04 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr24_-_25166416 | 2.04 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr8_-_32805214 | 2.02 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr13_+_16279890 | 1.98 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr19_+_4892281 | 1.98 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr6_+_39360377 | 1.96 |
ENSDART00000028260
ENSDART00000151322 |
zgc:77517
|
zgc:77517 |
| chr5_+_20428838 | 1.94 |
ENSDART00000141118
|
tmem119a
|
transmembrane protein 119a |
| chr25_+_25464630 | 1.92 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr6_+_4160579 | 1.89 |
ENSDART00000105278
ENSDART00000187932 ENSDART00000111817 |
trim25l
|
tripartite motif containing 25, like |
| chr3_-_32957702 | 1.89 |
ENSDART00000146586
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr12_+_34258139 | 1.89 |
ENSDART00000153127
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr23_+_40139765 | 1.89 |
ENSDART00000185376
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr22_-_17653143 | 1.87 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr17_-_19626357 | 1.86 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr3_+_30980852 | 1.84 |
ENSDART00000028529
|
prf1.1
|
perforin 1.1 |
| chr14_-_46198373 | 1.84 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr19_-_27777228 | 1.84 |
ENSDART00000046166
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr14_+_30774515 | 1.82 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr13_-_15994419 | 1.81 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr6_-_8580857 | 1.81 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr14_-_14566417 | 1.79 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr2_-_31634978 | 1.79 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr11_-_13126505 | 1.79 |
ENSDART00000158377
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr24_-_26304386 | 1.78 |
ENSDART00000175416
|
otos
|
otospiralin |
| chr1_+_14454663 | 1.76 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr8_-_25728628 | 1.75 |
ENSDART00000127237
|
foxp3a
|
forkhead box P3a |
| chr16_+_25196572 | 1.74 |
ENSDART00000141956
|
tyrobp
|
TYRO protein tyrosine kinase binding protein |
| chr14_-_33348221 | 1.74 |
ENSDART00000187749
|
rpl39
|
ribosomal protein L39 |
| chr1_+_11659861 | 1.74 |
ENSDART00000054787
|
BX569798.1
|
|
| chr2_-_29994726 | 1.73 |
ENSDART00000163350
|
cnpy1
|
canopy1 |
| chr24_+_25919809 | 1.73 |
ENSDART00000006615
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr9_-_1949915 | 1.71 |
ENSDART00000190712
|
hoxd3a
|
homeobox D3a |
| chr21_+_26657404 | 1.71 |
ENSDART00000129035
ENSDART00000186550 |
prdx5
|
peroxiredoxin 5 |
| chr6_+_54711306 | 1.70 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr14_-_1355544 | 1.70 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr3_-_34528306 | 1.70 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr6_+_30091811 | 1.67 |
ENSDART00000088403
|
meltf
|
melanotransferrin |
| chr14_+_30774894 | 1.66 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr19_-_27776649 | 1.65 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr11_+_34235372 | 1.64 |
ENSDART00000063150
|
fam43a
|
family with sequence similarity 43, member A |
| chr16_-_28878080 | 1.62 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr21_+_20356458 | 1.61 |
ENSDART00000135816
|
pde6b
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr12_-_32066469 | 1.61 |
ENSDART00000140685
ENSDART00000062185 |
rab40b
|
RAB40B, member RAS oncogene family |
| chr12_-_33789006 | 1.59 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr5_+_32882688 | 1.58 |
ENSDART00000008807
ENSDART00000185317 |
rpl12
|
ribosomal protein L12 |
| chr11_-_26040594 | 1.58 |
ENSDART00000144115
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr14_-_6666854 | 1.58 |
ENSDART00000133031
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr23_-_32162810 | 1.57 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr23_+_25428513 | 1.56 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr11_+_24716837 | 1.56 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr14_+_30774032 | 1.53 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr23_-_270847 | 1.50 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr13_+_25421531 | 1.50 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr13_-_35908275 | 1.49 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr9_-_32684008 | 1.49 |
ENSDART00000041751
|
ercc1
|
excision repair cross-complementation group 1 |
| chr7_+_19482084 | 1.48 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr2_-_58183499 | 1.46 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr11_+_43740949 | 1.45 |
ENSDART00000189296
|
CU862021.1
|
|
| chr1_+_40740951 | 1.43 |
ENSDART00000189780
ENSDART00000080563 |
htra3a
|
HtrA serine peptidase 3a |
| chr24_-_26622423 | 1.43 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr9_-_23994225 | 1.42 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr17_-_5352924 | 1.42 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr6_+_19950107 | 1.42 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr5_-_44286987 | 1.42 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr21_+_31253048 | 1.41 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr3_+_28576173 | 1.41 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr9_+_13985567 | 1.40 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr14_-_15956657 | 1.38 |
ENSDART00000169197
|
flt4
|
fms-related tyrosine kinase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 1.8 | 8.9 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 1.7 | 1.7 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) |
| 1.6 | 4.8 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 1.3 | 5.3 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.8 | 3.4 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.8 | 3.1 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.7 | 6.0 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.6 | 1.9 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.6 | 1.8 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.6 | 2.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.5 | 2.6 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.5 | 2.5 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.5 | 1.5 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.5 | 1.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.5 | 3.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.4 | 4.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 7.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.4 | 19.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.4 | 1.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.4 | 2.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 7.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.4 | 1.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.3 | 1.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 4.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 2.4 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.3 | 9.9 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.3 | 1.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 2.3 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.3 | 1.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.3 | 0.8 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.3 | 7.2 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.3 | 6.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.3 | 1.0 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.2 | 2.5 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.2 | 0.9 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.2 | 3.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 6.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 2.9 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.2 | 1.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 1.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 9.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 0.2 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 | 3.8 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 1.2 | GO:0031282 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 1.8 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.2 | 1.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 4.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.7 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 0.7 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 1.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 2.1 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.2 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 5.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.2 | 4.2 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 1.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.2 | 2.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 6.1 | GO:0035138 | pectoral fin morphogenesis(GO:0035138) |
| 0.2 | 4.9 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 3.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 5.0 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 0.6 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.6 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 6.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 8.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 1.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.7 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.7 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.8 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 4.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 3.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 5.0 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.1 | 2.8 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.4 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.8 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.3 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.4 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 6.8 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.1 | 3.3 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.1 | 5.5 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 1.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.8 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 2.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 1.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 1.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.3 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 0.2 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 3.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.1 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 2.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 3.3 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.1 | 0.3 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 2.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.0 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 2.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.7 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.9 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 1.9 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 3.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleotide catabolic process(GO:0009155) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 3.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.6 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 4.5 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.5 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 2.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 3.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 1.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 4.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.5 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 2.1 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 1.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 3.8 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.9 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.7 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.8 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.8 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.9 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.4 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.8 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.7 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 2.8 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 5.9 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 6.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 1.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 7.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 1.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 3.5 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.3 | 6.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.7 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.2 | 1.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 2.0 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 1.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.7 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.2 | 0.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 19.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.8 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.4 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 4.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 5.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 5.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.7 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 5.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.4 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 5.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 13.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 5.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 5.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 5.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 3.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.5 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 9.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 8.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 6.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 5.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.5 | 4.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.3 | 3.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.9 | 8.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.8 | 4.6 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.6 | 5.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.6 | 16.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 3.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 3.3 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 5.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 6.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.4 | 4.0 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 7.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 8.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 1.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 7.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 4.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 2.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 1.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.7 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.2 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 2.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 3.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.2 | 9.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 3.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 3.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 0.7 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.2 | 3.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 5.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 1.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 3.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.0 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 2.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 2.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.3 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 5.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 3.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 2.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 3.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 9.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.1 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 1.1 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 6.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 1.6 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 8.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 5.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 4.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 7.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 12.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 6.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 11.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.0 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 1.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 5.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 7.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.3 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 4.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 2.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 3.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 4.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 7.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.4 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 8.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 6.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.6 | 7.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.6 | 6.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.5 | 3.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 2.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 2.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 1.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 6.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 3.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 2.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 1.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 3.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 6.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 2.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.9 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.9 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.9 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.1 | 2.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 4.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.7 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |