Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
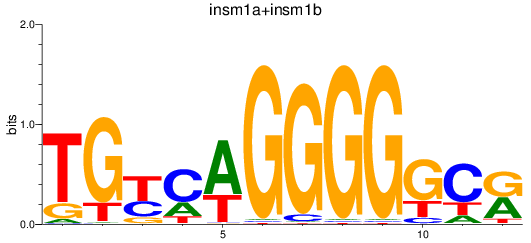
Results for insm1a+insm1b
Z-value: 0.50
Transcription factors associated with insm1a+insm1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
insm1b
|
ENSDARG00000053301 | insulinoma-associated 1b |
|
insm1a
|
ENSDARG00000091756 | insulinoma-associated 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| insm1b | dr11_v1_chr17_+_41463942_41463942 | 0.06 | 5.5e-01 | Click! |
| insm1a | dr11_v1_chr20_-_48485354_48485354 | 0.05 | 6.0e-01 | Click! |
Activity profile of insm1a+insm1b motif
Sorted Z-values of insm1a+insm1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_51830997 | 4.41 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr3_-_49566364 | 3.78 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr8_-_37391372 | 2.72 |
ENSDART00000190407
ENSDART00000009569 |
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr2_+_58221163 | 2.46 |
ENSDART00000157939
|
FO704813.1
|
|
| chr11_+_25619892 | 2.40 |
ENSDART00000146647
ENSDART00000033702 |
comtb
|
catechol-O-methyltransferase b |
| chr13_+_23176330 | 2.35 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr10_+_21801030 | 2.33 |
ENSDART00000160881
ENSDART00000187969 |
pcdh1g30
|
protocadherin 1 gamma 30 |
| chr8_-_14484599 | 2.30 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr17_-_4318393 | 2.26 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr7_+_23515966 | 2.25 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr4_+_13733838 | 2.10 |
ENSDART00000067166
ENSDART00000133157 |
cntn1b
|
contactin 1b |
| chr15_-_33925851 | 1.94 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr7_+_73397283 | 1.84 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr4_-_5597802 | 1.79 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr16_-_29458806 | 1.79 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr8_+_1082100 | 1.79 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr18_+_5137241 | 1.64 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr23_-_21797517 | 1.62 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr18_+_642889 | 1.60 |
ENSDART00000189007
|
CABZ01078320.1
|
|
| chr12_+_11352630 | 1.52 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr7_-_41489997 | 1.52 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr17_-_52579709 | 1.47 |
ENSDART00000156806
|
rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr3_-_6768905 | 1.42 |
ENSDART00000193638
ENSDART00000123809 ENSDART00000189440 ENSDART00000188335 |
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr7_-_72423666 | 1.36 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr19_+_46158078 | 1.35 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr9_-_4598883 | 1.34 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr14_-_6402769 | 1.29 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr12_-_4683325 | 1.28 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr9_-_1434484 | 1.27 |
ENSDART00000093412
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr18_+_50278858 | 1.26 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr18_+_48608366 | 1.26 |
ENSDART00000151229
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr11_+_24620742 | 1.25 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr17_+_45454943 | 1.24 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr9_+_53719230 | 1.24 |
ENSDART00000165827
|
PCDH8
|
si:ch211-199f5.1 |
| chr3_+_14388010 | 1.24 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr13_+_40034176 | 1.23 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr18_+_18612388 | 1.19 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr9_+_4429593 | 1.16 |
ENSDART00000184855
|
FP015810.1
|
|
| chr21_+_32332616 | 1.14 |
ENSDART00000154105
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr2_+_1202347 | 1.12 |
ENSDART00000075837
|
CABZ01084566.1
|
|
| chr7_+_73649686 | 1.11 |
ENSDART00000185589
|
PCP4L1
|
si:dkey-46i9.1 |
| chr11_-_30352333 | 1.11 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr17_+_9310259 | 1.11 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr10_-_41157135 | 1.10 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr3_+_17346502 | 1.08 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr4_+_69577362 | 1.08 |
ENSDART00000160913
|
znf1030
|
zinc finger protein 1030 |
| chr23_+_16430559 | 1.07 |
ENSDART00000112436
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr4_+_7888047 | 1.06 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr6_-_46875310 | 1.03 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr24_+_23959107 | 1.03 |
ENSDART00000180036
|
hgd
|
homogentisate 1,2-dioxygenase |
| chr19_+_9305964 | 1.00 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr24_-_33873451 | 0.99 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr12_-_48992527 | 0.98 |
ENSDART00000169696
|
CDHR1 (1 of many)
|
cadherin related family member 1 |
| chr16_-_31686602 | 0.95 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr22_-_881080 | 0.94 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr17_+_6793001 | 0.92 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr3_+_20001608 | 0.90 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr18_-_41211980 | 0.90 |
ENSDART00000171431
|
zbtb38
|
zinc finger and BTB domain containing 38 |
| chr10_+_41549819 | 0.89 |
ENSDART00000114210
|
CABZ01049847.1
|
|
| chr24_-_28306484 | 0.89 |
ENSDART00000148618
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr2_+_21486529 | 0.89 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr15_-_37921998 | 0.87 |
ENSDART00000193597
ENSDART00000181443 ENSDART00000168790 |
si:dkey-238d18.5
|
si:dkey-238d18.5 |
| chr5_-_10236599 | 0.86 |
ENSDART00000099834
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr8_-_49705058 | 0.85 |
ENSDART00000083760
|
frmd3
|
FERM domain containing 3 |
| chr22_+_15336752 | 0.85 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr4_+_40329524 | 0.85 |
ENSDART00000165722
|
znf993
|
zinc finger protein 993 |
| chr5_+_57641554 | 0.85 |
ENSDART00000171309
ENSDART00000157992 ENSDART00000164742 |
cryabb
|
crystallin, alpha B, b |
| chr9_-_29039506 | 0.82 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr14_-_4076480 | 0.81 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr9_+_38427572 | 0.80 |
ENSDART00000108860
|
CYP27A1 (1 of many)
|
zgc:136333 |
| chr14_+_8174828 | 0.80 |
ENSDART00000167228
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr9_-_42989297 | 0.80 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr12_+_47280839 | 0.79 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr10_+_44940693 | 0.79 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr14_-_28567845 | 0.76 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr19_+_1097393 | 0.75 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr9_-_41077934 | 0.73 |
ENSDART00000100342
|
ankar
|
ankyrin and armadillo repeat containing |
| chr15_-_47822597 | 0.72 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr20_+_1996202 | 0.71 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr7_-_72261721 | 0.69 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr2_+_9536021 | 0.68 |
ENSDART00000187357
|
CU929379.1
|
|
| chr23_+_19594608 | 0.67 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr9_-_30160897 | 0.65 |
ENSDART00000143986
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr16_-_22729491 | 0.63 |
ENSDART00000189854
|
tlr19
|
toll-like receptor 19 |
| chr6_-_39051319 | 0.63 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr21_+_21791799 | 0.63 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr12_+_47907277 | 0.62 |
ENSDART00000097714
|
tbata
|
thymus, brain and testes associated |
| chr13_-_49666615 | 0.62 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr19_-_19556778 | 0.61 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr25_-_28443607 | 0.61 |
ENSDART00000157243
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr13_+_18331509 | 0.60 |
ENSDART00000181918
|
zgc:110319
|
zgc:110319 |
| chr4_-_25908871 | 0.60 |
ENSDART00000066949
|
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr7_+_31871830 | 0.58 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr22_-_23666504 | 0.56 |
ENSDART00000158665
|
cfh
|
complement factor H |
| chr15_+_31471808 | 0.56 |
ENSDART00000110078
|
or102-3
|
odorant receptor, family C, subfamily 102, member 3 |
| chr10_+_6383270 | 0.56 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr4_-_61406364 | 0.56 |
ENSDART00000142592
|
znf1021
|
zinc finger protein 1021 |
| chr10_-_1180314 | 0.55 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr5_-_37252111 | 0.54 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr3_-_7129057 | 0.52 |
ENSDART00000125947
|
BX005085.1
|
|
| chr8_+_41453691 | 0.52 |
ENSDART00000061144
|
arpc5la
|
actin related protein 2/3 complex, subunit 5-like, a |
| chr8_+_23521974 | 0.51 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr16_+_38337783 | 0.51 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr2_-_16216568 | 0.51 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr14_+_8504410 | 0.50 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr11_+_15821433 | 0.50 |
ENSDART00000191005
|
FP003590.3
|
|
| chr25_-_35113891 | 0.48 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr23_+_12134839 | 0.46 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr5_+_50371951 | 0.46 |
ENSDART00000184488
ENSDART00000171295 |
slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr19_-_1411409 | 0.45 |
ENSDART00000159305
|
oxsm
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr18_+_40552620 | 0.44 |
ENSDART00000087699
|
si:ch211-132b12.1
|
si:ch211-132b12.1 |
| chr7_+_22585447 | 0.42 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr1_-_21599219 | 0.42 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr2_-_51316187 | 0.41 |
ENSDART00000163362
ENSDART00000166871 ENSDART00000160262 |
pigrl3.5
|
polymeric immunoglobulin receptor-like 3.5 |
| chr3_+_40672242 | 0.40 |
ENSDART00000188494
|
BX649559.2
|
|
| chr17_-_52609728 | 0.40 |
ENSDART00000103572
|
lrrc74a
|
leucine rich repeat containing 74A |
| chr21_+_25660613 | 0.40 |
ENSDART00000134017
|
si:dkey-17e16.15
|
si:dkey-17e16.15 |
| chr3_-_40051425 | 0.39 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr12_+_30368145 | 0.38 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr1_-_40341306 | 0.38 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr7_-_3470451 | 0.38 |
ENSDART00000173387
|
si:ch211-285c6.6
|
si:ch211-285c6.6 |
| chr2_-_51454868 | 0.37 |
ENSDART00000172550
|
si:dkeyp-104b3.2
|
si:dkeyp-104b3.2 |
| chr2_-_28682476 | 0.36 |
ENSDART00000190178
|
dhcr7
|
7-dehydrocholesterol reductase |
| chr10_-_40352250 | 0.35 |
ENSDART00000150821
|
taar20f
|
trace amine associated receptor 20f |
| chr1_+_2112726 | 0.34 |
ENSDART00000131714
ENSDART00000138396 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr6_-_55354004 | 0.34 |
ENSDART00000165911
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr12_-_47774807 | 0.34 |
ENSDART00000193831
|
LO017725.1
|
|
| chr13_+_36680564 | 0.31 |
ENSDART00000136030
ENSDART00000006923 |
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex subunit s |
| chr11_-_20956309 | 0.31 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr21_+_26612777 | 0.31 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr23_-_22623115 | 0.30 |
ENSDART00000188049
|
CR388207.3
|
|
| chr13_-_36680531 | 0.29 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr1_-_18803919 | 0.29 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr10_+_14210667 | 0.29 |
ENSDART00000138854
|
si:dkey-286h21.1
|
si:dkey-286h21.1 |
| chr3_-_61592417 | 0.29 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr3_+_55131289 | 0.28 |
ENSDART00000111585
|
AL935210.1
|
|
| chr6_-_10828880 | 0.28 |
ENSDART00000131458
ENSDART00000020261 |
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr9_+_29040425 | 0.28 |
ENSDART00000150201
|
MRAS
|
si:ch73-116o1.2 |
| chr23_-_25891426 | 0.28 |
ENSDART00000189907
|
CR318614.1
|
|
| chr18_+_17493859 | 0.26 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr3_+_55114097 | 0.26 |
ENSDART00000121686
|
hbbe1.1
|
hemoglobin beta embryonic-1.1 |
| chr21_+_40261187 | 0.26 |
ENSDART00000142204
|
si:ch211-218m3.11
|
si:ch211-218m3.11 |
| chr25_-_25575241 | 0.24 |
ENSDART00000150636
|
hic1l
|
hypermethylated in cancer 1 like |
| chr25_-_13659249 | 0.22 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr11_-_42099645 | 0.22 |
ENSDART00000173312
|
abhd6a
|
abhydrolase domain containing 6a |
| chr25_+_16601839 | 0.21 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr1_+_12767318 | 0.20 |
ENSDART00000162652
|
pcdh10a
|
protocadherin 10a |
| chr3_+_18437397 | 0.20 |
ENSDART00000136243
ENSDART00000184882 ENSDART00000135470 |
tbc1d16
|
TBC1 domain family, member 16 |
| chr1_+_56955349 | 0.20 |
ENSDART00000152758
|
si:ch211-1f22.10
|
si:ch211-1f22.10 |
| chr7_-_17412559 | 0.19 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr13_+_41819817 | 0.18 |
ENSDART00000185778
|
CABZ01066611.1
|
|
| chr11_-_309420 | 0.18 |
ENSDART00000173185
|
poc1a
|
POC1 centriolar protein A |
| chr13_-_50546634 | 0.18 |
ENSDART00000192127
|
CU570781.2
|
|
| chr13_+_13668991 | 0.17 |
ENSDART00000148266
|
pimr48
|
Pim proto-oncogene, serine/threonine kinase, related 48 |
| chr2_-_37098785 | 0.16 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr22_+_344763 | 0.15 |
ENSDART00000181934
|
CU914780.1
|
|
| chr3_+_55122662 | 0.15 |
ENSDART00000128380
|
hbbe1.2
|
hemoglobin beta embryonic-1.2 |
| chr14_+_42172677 | 0.13 |
ENSDART00000148544
|
pcdh18b
|
protocadherin 18b |
| chr8_+_15251448 | 0.12 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr21_-_30415524 | 0.12 |
ENSDART00000101036
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr19_-_24198139 | 0.11 |
ENSDART00000138864
ENSDART00000125350 |
prf1.7
|
perforin 1.7 |
| chr25_-_19661198 | 0.11 |
ENSDART00000149641
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr5_-_16274058 | 0.11 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr25_-_21085661 | 0.11 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr6_+_9421279 | 0.10 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr25_-_24046870 | 0.10 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr25_-_25575717 | 0.10 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr17_+_25197180 | 0.10 |
ENSDART00000148775
|
arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr5_-_37044262 | 0.07 |
ENSDART00000166670
|
si:dkeyp-110c7.8
|
si:dkeyp-110c7.8 |
| chr1_+_55452892 | 0.07 |
ENSDART00000122508
|
CR788255.1
|
|
| chr19_+_9032073 | 0.06 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr15_+_4988189 | 0.04 |
ENSDART00000142995
ENSDART00000062852 |
spcs2
|
signal peptidase complex subunit 2 |
| chr5_+_36932718 | 0.03 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr4_+_76466751 | 0.03 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr3_+_12829566 | 0.02 |
ENSDART00000157672
|
si:ch211-8c17.2
|
si:ch211-8c17.2 |
| chr24_-_10394277 | 0.01 |
ENSDART00000127568
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr4_-_17409533 | 0.00 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
Network of associatons between targets according to the STRING database.
First level regulatory network of insm1a+insm1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.6 | 2.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 2.4 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.3 | 0.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 2.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 2.3 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 1.1 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.5 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 2.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 2.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.5 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 1.4 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.1 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 1.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.3 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.0 | 1.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.5 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.9 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.4 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.1 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.9 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 2.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.6 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 6.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.6 | 2.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.5 | 1.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 1.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 1.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.4 | 2.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 1.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 2.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.9 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 2.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 2.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.3 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 1.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.6 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 1.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 1.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |