Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
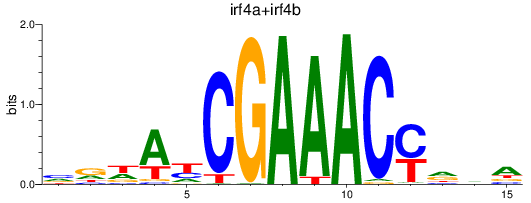
Results for irf4a+irf4b
Z-value: 1.14
Transcription factors associated with irf4a+irf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf4a
|
ENSDARG00000006560 | interferon regulatory factor 4a |
|
irf4b
|
ENSDARG00000055374 | interferon regulatory factor 4b |
|
irf4b
|
ENSDARG00000115381 | interferon regulatory factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf4b | dr11_v1_chr20_+_26556174_26556174 | 0.59 | 3.8e-10 | Click! |
| irf4a | dr11_v1_chr2_-_879800_879800 | 0.20 | 5.7e-02 | Click! |
Activity profile of irf4a+irf4b motif
Sorted Z-values of irf4a+irf4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_22552678 | 14.35 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr19_-_7110617 | 12.77 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr4_-_17669881 | 11.11 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr7_+_19482877 | 9.54 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr19_-_15420678 | 9.50 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr7_+_19483277 | 9.02 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr16_+_19637384 | 8.68 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr9_-_32837860 | 8.61 |
ENSDART00000142227
|
mxe
|
myxovirus (influenza virus) resistance E |
| chr16_+_48753664 | 8.47 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr4_-_12795436 | 8.43 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr9_+_23765587 | 8.35 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr16_+_29492937 | 8.17 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr2_-_37140423 | 8.06 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr23_-_30045661 | 7.66 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr4_-_26107841 | 7.46 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr15_-_40175894 | 7.06 |
ENSDART00000156632
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr4_-_12795030 | 6.91 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr1_-_7570181 | 6.62 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr14_-_33585809 | 6.41 |
ENSDART00000023540
|
sash3
|
SAM and SH3 domain containing 3 |
| chr16_+_46497149 | 6.06 |
ENSDART00000135151
ENSDART00000058324 |
rpz4
|
rapunzel 4 |
| chr19_+_791538 | 5.95 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr7_-_34192834 | 5.94 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr21_+_45626136 | 5.83 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr15_+_34069746 | 5.79 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr12_-_23658888 | 5.79 |
ENSDART00000088319
|
map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr24_-_11467549 | 5.78 |
ENSDART00000082264
|
pxdc1b
|
PX domain containing 1b |
| chr10_+_8101729 | 5.77 |
ENSDART00000138875
ENSDART00000123447 ENSDART00000185333 |
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr7_-_26579465 | 5.73 |
ENSDART00000173820
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr11_+_10984293 | 5.67 |
ENSDART00000065933
|
itgb6
|
integrin, beta 6 |
| chr7_-_19614916 | 5.56 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr14_+_33329761 | 5.56 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr1_+_38153944 | 5.45 |
ENSDART00000135666
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr1_+_31674297 | 5.40 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr8_-_27687095 | 5.32 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr14_+_33329420 | 5.23 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr9_-_23765480 | 5.03 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr24_-_11467004 | 4.93 |
ENSDART00000144316
|
pxdc1b
|
PX domain containing 1b |
| chr3_-_29941357 | 4.72 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr22_-_17474583 | 4.37 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr3_+_25191467 | 4.37 |
ENSDART00000156956
ENSDART00000154799 |
il2rb
|
interleukin 2 receptor, beta |
| chr22_-_17474781 | 4.23 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr7_+_19615056 | 4.17 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr21_+_25793970 | 3.97 |
ENSDART00000101217
|
CLDN4 (1 of many)
|
zgc:136892 |
| chr18_+_44532370 | 3.92 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr2_-_13333932 | 3.89 |
ENSDART00000150238
ENSDART00000168258 |
si:dkey-185p13.1
vps4b
|
si:dkey-185p13.1 vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr4_+_11464255 | 3.84 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr25_-_774350 | 3.83 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr18_+_17624295 | 3.79 |
ENSDART00000151865
ENSDART00000190408 ENSDART00000189204 ENSDART00000188894 ENSDART00000033762 ENSDART00000193471 |
nlrc5
|
NLR family, CARD domain containing 5 |
| chr15_-_41734639 | 3.75 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr13_+_24263049 | 3.74 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr9_+_42269059 | 3.51 |
ENSDART00000113435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr5_-_69716501 | 3.50 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr21_-_37933833 | 3.44 |
ENSDART00000184138
ENSDART00000177664 |
FO704750.1
|
|
| chr4_+_68562464 | 3.40 |
ENSDART00000192954
|
BX548011.4
|
|
| chr22_-_30999000 | 3.38 |
ENSDART00000109841
|
ssuh2.3
|
ssu-2 homolog, tandem duplicate 3 |
| chr18_+_44532668 | 3.31 |
ENSDART00000140672
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr10_+_5060191 | 3.22 |
ENSDART00000145908
ENSDART00000122397 |
carm1l
|
coactivator-associated arginine methyltransferase 1, like |
| chr12_-_35936329 | 3.18 |
ENSDART00000166634
|
rnf213b
|
ring finger protein 213b |
| chr25_+_16646113 | 3.14 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr4_-_6373735 | 3.14 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr15_-_17071328 | 3.13 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr25_+_28862660 | 2.86 |
ENSDART00000154681
|
si:ch211-106e7.2
|
si:ch211-106e7.2 |
| chr23_-_29553430 | 2.84 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr25_+_15997957 | 2.78 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr17_+_35362851 | 2.75 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr18_+_44532199 | 2.66 |
ENSDART00000135386
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr2_+_2818645 | 2.56 |
ENSDART00000163587
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr18_+_44532883 | 2.56 |
ENSDART00000121994
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr11_-_42472941 | 2.54 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr2_-_24554416 | 2.49 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr25_-_3034668 | 2.47 |
ENSDART00000053405
|
scamp2
|
secretory carrier membrane protein 2 |
| chr2_-_55797318 | 2.32 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr6_-_18400548 | 2.31 |
ENSDART00000179797
ENSDART00000164891 |
trim25
|
tripartite motif containing 25 |
| chr7_-_20464133 | 2.29 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr1_-_35924495 | 2.29 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr8_+_45361775 | 2.20 |
ENSDART00000015193
|
chmp4bb
|
charged multivesicular body protein 4Bb |
| chr9_+_34089156 | 2.19 |
ENSDART00000000005
|
ccdc80
|
coiled-coil domain containing 80 |
| chr4_-_18434924 | 2.11 |
ENSDART00000190271
|
socs2
|
suppressor of cytokine signaling 2 |
| chr25_-_483808 | 2.05 |
ENSDART00000104720
|
si:ch1073-385f13.3
|
si:ch1073-385f13.3 |
| chr10_+_28160265 | 2.03 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr15_+_2421729 | 1.97 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr5_+_21922534 | 1.95 |
ENSDART00000148092
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr10_+_4046448 | 1.89 |
ENSDART00000123086
ENSDART00000052268 |
pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr13_+_22476742 | 1.85 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr9_+_23003208 | 1.82 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr20_-_3911546 | 1.82 |
ENSDART00000169787
|
cnksr3
|
cnksr family member 3 |
| chr20_-_51547464 | 1.81 |
ENSDART00000099486
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr20_-_25436389 | 1.79 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr15_-_41749364 | 1.73 |
ENSDART00000155464
|
ftr73
|
finTRIM family, member 73 |
| chr3_+_36127287 | 1.60 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr15_+_2421432 | 1.60 |
ENSDART00000193772
|
hephl1a
|
hephaestin-like 1a |
| chr20_-_35012093 | 1.53 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr13_-_50200348 | 1.52 |
ENSDART00000038391
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr16_+_27383717 | 1.50 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr24_-_26885897 | 1.46 |
ENSDART00000180512
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr8_-_51340773 | 1.45 |
ENSDART00000060633
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr19_-_19871211 | 1.45 |
ENSDART00000170980
|
evx1
|
even-skipped homeobox 1 |
| chr1_-_55750208 | 1.38 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr18_-_24988645 | 1.33 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr10_+_7703251 | 1.29 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr7_+_17908235 | 1.24 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr22_+_19365220 | 1.22 |
ENSDART00000132781
ENSDART00000135672 ENSDART00000153630 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr6_+_22068589 | 1.21 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr15_-_43995028 | 1.19 |
ENSDART00000172485
ENSDART00000186320 |
nlrc3l
|
NLR family, CARD domain containing 3-like |
| chr1_-_714626 | 1.18 |
ENSDART00000161072
|
adamts5
|
ADAM metallopeptidase with thrombospondin type 1 motif 5 |
| chr23_-_29553691 | 1.04 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr12_+_48319221 | 0.99 |
ENSDART00000126362
|
anapc16
|
anaphase promoting complex subunit 16 |
| chr21_-_40049642 | 0.90 |
ENSDART00000124416
|
med31
|
mediator complex subunit 31 |
| chr2_-_43739740 | 0.87 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr13_-_50200042 | 0.87 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr15_-_21702317 | 0.84 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr5_+_66063807 | 0.82 |
ENSDART00000122405
|
jak2b
|
Janus kinase 2b |
| chr12_-_28975705 | 0.79 |
ENSDART00000129173
|
znf646
|
zinc finger protein 646 |
| chr22_-_8692305 | 0.79 |
ENSDART00000181602
|
CR450686.4
|
|
| chr24_-_3407507 | 0.76 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr2_-_26620803 | 0.74 |
ENSDART00000003946
ENSDART00000126826 |
tmem59
|
transmembrane protein 59 |
| chr15_-_29556757 | 0.59 |
ENSDART00000060049
|
hspa13
|
heat shock protein 70 family, member 13 |
| chr3_-_4501026 | 0.54 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr17_-_23603263 | 0.50 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr10_-_5821584 | 0.50 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr21_-_22543611 | 0.45 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr22_+_38914983 | 0.44 |
ENSDART00000085701
|
senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr25_-_37489917 | 0.42 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr19_-_791016 | 0.39 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr19_-_42462491 | 0.39 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr5_+_35502290 | 0.36 |
ENSDART00000191222
|
CR762390.1
|
|
| chr9_+_51147764 | 0.33 |
ENSDART00000188480
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr3_-_26171668 | 0.24 |
ENSDART00000113890
|
rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr5_+_31959954 | 0.23 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr3_-_49554912 | 0.18 |
ENSDART00000159392
|
CR847534.1
|
|
| chr18_-_23874929 | 0.16 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_15586632 | 0.12 |
ENSDART00000164903
|
BX510342.1
|
|
| chr11_-_30508843 | 0.12 |
ENSDART00000101667
ENSDART00000179930 |
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr5_+_28260158 | 0.09 |
ENSDART00000181434
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr20_+_13781779 | 0.06 |
ENSDART00000142999
ENSDART00000152471 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr8_+_38564942 | 0.06 |
ENSDART00000085371
|
rfk
|
riboflavin kinase |
| chr20_-_38525467 | 0.05 |
ENSDART00000061417
|
si:ch211-245h14.1
|
si:ch211-245h14.1 |
| chr1_-_7582859 | 0.04 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr15_-_1835189 | 0.00 |
ENSDART00000154369
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf4a+irf4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 19.1 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 1.1 | 5.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.9 | 2.8 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.9 | 2.6 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.8 | 5.9 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.8 | 6.6 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.8 | 8.1 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.7 | 2.2 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.6 | 3.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.6 | 4.7 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.5 | 1.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.5 | 8.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.4 | 13.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 2.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.4 | 2.0 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.4 | 1.2 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 1.2 | GO:0014005 | microglia development(GO:0014005) |
| 0.2 | 1.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 5.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 5.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 3.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 0.5 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.2 | 1.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 2.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 1.8 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 0.9 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.8 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 3.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 0.4 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 8.2 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 2.5 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 2.3 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 12.4 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 14.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 4.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.5 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 11.1 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 2.2 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 2.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 13.2 | GO:0072594 | establishment of protein localization to organelle(GO:0072594) |
| 0.1 | 2.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 3.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 3.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 1.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 8.7 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 5.3 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 2.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 5.9 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 4.6 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 6.5 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 5.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 8.3 | GO:0006886 | intracellular protein transport(GO:0006886) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 1.8 | GO:0016197 | endosomal transport(GO:0016197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 14.4 | GO:0043034 | costamere(GO:0043034) |
| 2.6 | 15.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.9 | 5.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.8 | 15.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.5 | 13.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.4 | 8.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 8.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 20.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.3 | 3.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 1.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 5.3 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 5.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 20.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 7.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 3.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 5.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 5.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 3.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 1.3 | 3.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.9 | 2.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.7 | 13.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.6 | 2.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.6 | 3.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.5 | 5.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.5 | 3.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.4 | 8.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.4 | 1.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.4 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 3.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 5.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 1.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.3 | 2.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.3 | 1.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.3 | 1.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 1.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 2.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 5.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 3.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 4.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 5.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.8 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 13.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 8.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 10.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 4.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.3 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 6.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 10.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 14.0 | GO:0004672 | protein kinase activity(GO:0004672) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 17.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.5 | 5.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 2.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 3.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 2.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 3.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.9 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 2.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 7.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 3.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 3.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.7 | 8.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.5 | 3.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 5.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 4.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 6.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 3.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 5.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 5.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.9 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |