Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
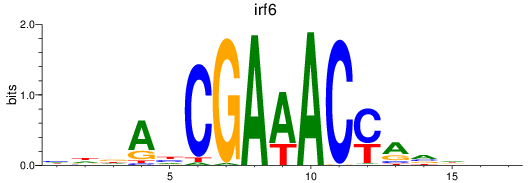
Results for irf6
Z-value: 0.50
Transcription factors associated with irf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf6
|
ENSDARG00000101986 | interferon regulatory factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf6 | dr11_v1_chr22_+_1170294_1170295 | 0.68 | 6.8e-14 | Click! |
Activity profile of irf6 motif
Sorted Z-values of irf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_22552678 | 10.11 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr16_+_19637384 | 5.36 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr4_-_17669881 | 4.48 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr19_-_15420678 | 4.47 |
ENSDART00000151454
ENSDART00000027697 |
serinc2
|
serine incorporator 2 |
| chr18_+_44532370 | 4.19 |
ENSDART00000086952
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr7_+_69187585 | 4.12 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr17_-_30666037 | 3.34 |
ENSDART00000156509
|
alkal2b
|
ALK and LTK ligand 2b |
| chr18_+_44532668 | 3.06 |
ENSDART00000140672
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr14_+_33329420 | 3.03 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr18_+_44532199 | 3.02 |
ENSDART00000135386
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr7_+_19482877 | 2.92 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr14_+_33329761 | 2.91 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr18_+_44532883 | 2.75 |
ENSDART00000121994
|
st14a
|
suppression of tumorigenicity 14 (colon carcinoma) a |
| chr2_-_37140423 | 2.73 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr9_+_23765587 | 2.34 |
ENSDART00000145120
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr7_+_19483277 | 1.75 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr18_+_17624295 | 1.70 |
ENSDART00000151865
ENSDART00000190408 ENSDART00000189204 ENSDART00000188894 ENSDART00000033762 ENSDART00000193471 |
nlrc5
|
NLR family, CARD domain containing 5 |
| chr23_-_30045661 | 1.67 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr9_-_23765480 | 1.54 |
ENSDART00000027212
|
si:ch211-219a4.6
|
si:ch211-219a4.6 |
| chr20_-_3911546 | 1.21 |
ENSDART00000169787
|
cnksr3
|
cnksr family member 3 |
| chr21_-_22543611 | 1.13 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr16_+_27383717 | 0.88 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr8_-_27687095 | 0.68 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr10_+_28160265 | 0.64 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr15_-_21702317 | 0.54 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr2_-_26620803 | 0.43 |
ENSDART00000003946
ENSDART00000126826 |
tmem59
|
transmembrane protein 59 |
| chr9_+_34089156 | 0.41 |
ENSDART00000000005
|
ccdc80
|
coiled-coil domain containing 80 |
| chr16_+_26601364 | 0.08 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.6 | 3.3 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.3 | 1.7 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.3 | 1.1 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.7 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.6 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 13.0 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 10.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 4.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 5.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 3.3 | GO:0072594 | establishment of protein localization to organelle(GO:0072594) |
| 0.0 | 0.4 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.1 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 4.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 4.9 | GO:0005764 | lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.2 | 0.7 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 13.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |