Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
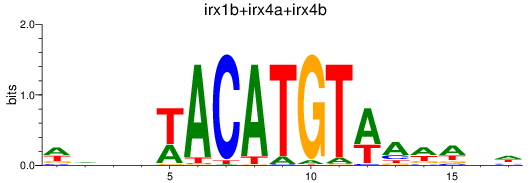
Results for irx1b+irx4a+irx4b
Z-value: 0.50
Transcription factors associated with irx1b+irx4a+irx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx4a
|
ENSDARG00000035648 | iroquois homeobox 4a |
|
irx4b
|
ENSDARG00000036051 | iroquois homeobox 4b |
|
irx1b
|
ENSDARG00000056594 | iroquois homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx1b | dr11_v1_chr19_-_28130658_28130658 | -0.70 | 2.9e-15 | Click! |
| irx4b | dr11_v1_chr19_+_28116410_28116410 | -0.53 | 4.8e-08 | Click! |
| irx4a | dr11_v1_chr16_-_383664_383664 | -0.47 | 2.0e-06 | Click! |
Activity profile of irx1b+irx4a+irx4b motif
Sorted Z-values of irx1b+irx4a+irx4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50147948 | 10.46 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr20_+_23440632 | 9.90 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr5_+_66170479 | 7.45 |
ENSDART00000172117
|
gldc
|
glycine dehydrogenase (decarboxylating) |
| chr13_+_2908764 | 5.92 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr23_+_19701587 | 5.38 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr7_+_56577522 | 5.13 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr7_+_56577906 | 4.28 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr18_+_22109379 | 3.76 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr1_-_7951002 | 2.93 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr24_-_25098719 | 2.87 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr5_-_35159379 | 2.42 |
ENSDART00000144105
|
fcho2
|
FCH domain only 2 |
| chr3_-_402714 | 2.30 |
ENSDART00000134062
ENSDART00000105659 |
mhc1zja
|
major histocompatibility complex class I ZJA |
| chr11_+_14284866 | 2.18 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr13_-_43149063 | 2.04 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr20_-_23440955 | 1.93 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr25_-_29072162 | 1.91 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr3_-_34100700 | 1.84 |
ENSDART00000151628
|
ighv6-1
|
immunoglobulin heavy variable 6-1 |
| chr21_-_45871866 | 1.64 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr20_-_27190393 | 1.56 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr6_-_43262400 | 1.40 |
ENSDART00000156267
|
frmd4ba
|
FERM domain containing 4Ba |
| chr16_-_45398408 | 1.20 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr19_+_3653976 | 1.19 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr8_+_26410197 | 1.14 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr17_-_5352924 | 1.14 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr23_+_40275400 | 1.05 |
ENSDART00000184259
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr15_+_37331585 | 1.02 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr2_-_7185460 | 0.98 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr2_-_13333932 | 0.82 |
ENSDART00000150238
ENSDART00000168258 |
si:dkey-185p13.1
vps4b
|
si:dkey-185p13.1 vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr10_+_45345574 | 0.80 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr1_+_53919110 | 0.76 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr16_-_44709832 | 0.65 |
ENSDART00000156784
|
si:ch211-151m7.6
|
si:ch211-151m7.6 |
| chr15_-_41734639 | 0.62 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr2_-_707152 | 0.58 |
ENSDART00000082304
|
ftr93
|
finTRIM family, member 93 |
| chr5_-_58996324 | 0.51 |
ENSDART00000033923
|
mis12
|
MIS12 kinetochore complex component |
| chr10_-_22912255 | 0.42 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
| chr8_+_29986265 | 0.38 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr10_-_28117740 | 0.38 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr23_+_45845423 | 0.36 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr14_-_10617127 | 0.35 |
ENSDART00000154299
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr7_-_31830936 | 0.31 |
ENSDART00000052514
ENSDART00000129720 |
cars
|
cysteinyl-tRNA synthetase |
| chr8_+_23784471 | 0.27 |
ENSDART00000189457
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr23_+_45845159 | 0.26 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr23_+_40275601 | 0.26 |
ENSDART00000076876
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr21_+_12036238 | 0.10 |
ENSDART00000102463
ENSDART00000155426 |
zgc:162344
|
zgc:162344 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irx1b+irx4a+irx4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.5 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 | 5.4 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.2 | 1.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.8 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.4 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 2.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.5 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.4 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 1.6 | GO:0061138 | morphogenesis of a branching epithelium(GO:0061138) |
| 0.0 | 1.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.0 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 2.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 19.3 | GO:0006508 | proteolysis(GO:0006508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.2 | 1.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 2.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 20.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.5 | GO:0016594 | glycine binding(GO:0016594) |
| 0.5 | 5.4 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 20.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 2.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |