Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for irx2a
Z-value: 0.83
Transcription factors associated with irx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx2a
|
ENSDARG00000001785 | iroquois homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx2a | dr11_v1_chr16_-_563235_563235 | 0.66 | 6.2e-13 | Click! |
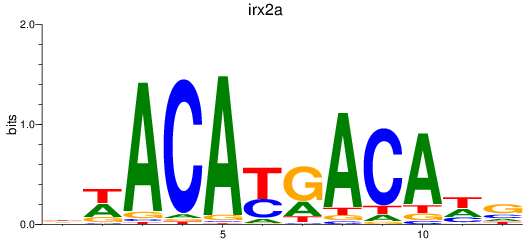
Activity profile of irx2a motif
Sorted Z-values of irx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_31282161 | 12.77 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr6_+_27146671 | 10.83 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr24_-_7632187 | 8.52 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr2_+_47581997 | 8.07 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr2_-_13216269 | 7.26 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr6_-_54845622 | 6.93 |
ENSDART00000048887
|
csrp1b
|
cysteine and glycine-rich protein 1b |
| chr17_-_20897250 | 6.66 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr5_-_4493502 | 6.50 |
ENSDART00000067593
ENSDART00000190221 |
zgc:153704
|
zgc:153704 |
| chr7_+_23875269 | 6.47 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr19_+_30387999 | 6.35 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr2_+_47582681 | 6.16 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr12_+_9761685 | 5.86 |
ENSDART00000189522
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr16_-_43025885 | 5.79 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr3_+_54849083 | 5.79 |
ENSDART00000008988
ENSDART00000078221 |
mgrn1b
|
mahogunin, ring finger 1b |
| chr12_-_10275320 | 5.76 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr6_-_54845083 | 5.59 |
ENSDART00000148815
|
csrp1b
|
cysteine and glycine-rich protein 1b |
| chr2_+_22795494 | 5.43 |
ENSDART00000042255
|
rab6bb
|
RAB6B, member RAS oncogene family b |
| chr24_+_14713776 | 5.36 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr23_-_28141419 | 5.19 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr1_+_20635190 | 5.10 |
ENSDART00000145418
ENSDART00000148518 ENSDART00000139461 ENSDART00000102969 ENSDART00000166479 |
spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr16_+_34479064 | 5.09 |
ENSDART00000159965
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr8_+_25959940 | 5.09 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr7_-_18656069 | 5.07 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr19_+_30388186 | 4.84 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr9_-_32730487 | 4.69 |
ENSDART00000147795
|
olig1
|
oligodendrocyte transcription factor 1 |
| chr5_-_22621417 | 4.67 |
ENSDART00000138716
|
zgc:113208
|
zgc:113208 |
| chr2_-_36932253 | 4.50 |
ENSDART00000124217
|
map1sb
|
microtubule-associated protein 1Sb |
| chr6_+_12326267 | 4.43 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr5_+_31214341 | 4.40 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr12_+_26632448 | 4.33 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr6_-_18698006 | 4.24 |
ENSDART00000170128
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr2_+_38924975 | 4.22 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr17_-_35881841 | 4.21 |
ENSDART00000110040
|
sox11a
|
SRY (sex determining region Y)-box 11a |
| chr5_+_21931124 | 4.05 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr23_+_4299887 | 3.94 |
ENSDART00000132604
|
l3mbtl1a
|
l(3)mbt-like 1a (Drosophila) |
| chr7_-_34339845 | 3.92 |
ENSDART00000173816
|
madd
|
MAP-kinase activating death domain |
| chr23_-_39849155 | 3.91 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr11_-_21585176 | 3.75 |
ENSDART00000045391
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr20_+_38201644 | 3.74 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr1_+_41666611 | 3.68 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr20_+_26349002 | 3.59 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr8_-_25011157 | 3.55 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr3_-_33494163 | 3.54 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr22_-_28226948 | 3.52 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr3_-_43821381 | 3.52 |
ENSDART00000166021
|
snn
|
stannin |
| chr8_+_23147218 | 3.49 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr4_+_8168514 | 3.48 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr16_-_52821023 | 3.01 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr7_-_5431841 | 2.99 |
ENSDART00000173073
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_+_28831450 | 2.90 |
ENSDART00000055422
|
flr
|
fleer |
| chr21_-_2248239 | 2.87 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr9_+_41024973 | 2.87 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr1_+_52518176 | 2.87 |
ENSDART00000003278
|
tacr3l
|
tachykinin receptor 3-like |
| chr25_-_36286186 | 2.85 |
ENSDART00000182398
|
slc7a10b
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10b |
| chr14_+_32022272 | 2.83 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr16_-_22022456 | 2.82 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| chr12_+_27024676 | 2.76 |
ENSDART00000153104
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr21_-_4032650 | 2.75 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr21_+_21339311 | 2.71 |
ENSDART00000137693
ENSDART00000161820 |
sdhaf1
si:ch211-191j22.3
|
succinate dehydrogenase complex assembly factor 1 si:ch211-191j22.3 |
| chr19_+_34311374 | 2.67 |
ENSDART00000086617
|
gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr1_+_15226268 | 2.66 |
ENSDART00000109911
|
hgsnat
|
heparan-alpha-glucosaminide N-acetyltransferase |
| chr2_+_47582488 | 2.66 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr21_+_723998 | 2.64 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr19_-_42416696 | 2.63 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr6_+_28205284 | 2.62 |
ENSDART00000160707
ENSDART00000190509 |
smx5
LSM2 (1 of many)
|
smx5 si:ch73-14h10.2 |
| chr22_+_2715815 | 2.58 |
ENSDART00000042770
|
znf1163
|
zinc finger protein 1163 |
| chr2_-_42864472 | 2.57 |
ENSDART00000134139
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr11_+_38332419 | 2.55 |
ENSDART00000005864
|
CDK18
|
si:dkey-166c18.1 |
| chr2_-_4797512 | 2.53 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr21_-_32467799 | 2.52 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr19_+_7549854 | 2.52 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr16_-_25393699 | 2.48 |
ENSDART00000149445
|
pard6ga
|
par-6 family cell polarity regulator gamma a |
| chr19_+_42735211 | 2.45 |
ENSDART00000139109
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr23_+_23658474 | 2.43 |
ENSDART00000162838
|
agrn
|
agrin |
| chr21_-_4244514 | 2.37 |
ENSDART00000065880
|
uck1
|
uridine-cytidine kinase 1 |
| chr18_-_29961784 | 2.36 |
ENSDART00000135601
ENSDART00000140661 |
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr22_-_354592 | 2.36 |
ENSDART00000155769
|
tmem240b
|
transmembrane protein 240b |
| chr19_+_10832092 | 2.34 |
ENSDART00000191851
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr6_-_19340889 | 2.33 |
ENSDART00000181407
|
mif4gda
|
MIF4G domain containing a |
| chr24_-_37955993 | 2.33 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr17_-_20897407 | 2.31 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr15_+_5321612 | 2.27 |
ENSDART00000174345
|
or113-1
|
odorant receptor, family A, subfamily 113, member 1 |
| chr8_-_25566347 | 2.24 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr19_-_41069573 | 2.17 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr7_+_50109239 | 2.16 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr1_-_11878845 | 2.15 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr5_-_12560569 | 2.10 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr21_+_5915041 | 2.10 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr9_+_54417141 | 2.04 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr6_-_10776254 | 2.04 |
ENSDART00000184008
|
gpr155b
|
G protein-coupled receptor 155b |
| chr25_-_13676682 | 2.03 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr21_+_6961635 | 1.98 |
ENSDART00000137281
|
si:ch211-93g21.2
|
si:ch211-93g21.2 |
| chr2_-_44038698 | 1.96 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr11_+_12811906 | 1.95 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr16_+_24681177 | 1.95 |
ENSDART00000058956
ENSDART00000189335 |
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr7_+_13582256 | 1.95 |
ENSDART00000158477
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr5_+_36650096 | 1.94 |
ENSDART00000111414
|
alkbh6
|
alkB homolog 6 |
| chr21_-_2238277 | 1.93 |
ENSDART00000165754
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr5_+_31860043 | 1.91 |
ENSDART00000036235
ENSDART00000140541 |
iscub
|
iron-sulfur cluster assembly enzyme b |
| chr20_-_33790003 | 1.91 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr17_+_32571584 | 1.88 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr14_-_25985698 | 1.88 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr2_+_30739680 | 1.88 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr13_-_24825691 | 1.87 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr7_+_39624728 | 1.87 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr22_-_15010688 | 1.87 |
ENSDART00000139892
|
elfn2a
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2a |
| chr22_+_21972824 | 1.83 |
ENSDART00000183708
ENSDART00000133739 |
gna15.3
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 |
| chr25_+_7671640 | 1.82 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr2_-_10014821 | 1.80 |
ENSDART00000185525
|
CR956623.1
|
|
| chr2_+_30960351 | 1.79 |
ENSDART00000141575
|
lpin2
|
lipin 2 |
| chr4_+_77060861 | 1.77 |
ENSDART00000174271
ENSDART00000174393 ENSDART00000150450 |
si:dkey-240n22.8
|
si:dkey-240n22.8 |
| chr6_-_18698456 | 1.75 |
ENSDART00000166587
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr21_-_2232640 | 1.74 |
ENSDART00000157754
|
zgc:113343
|
zgc:113343 |
| chr16_+_32131568 | 1.73 |
ENSDART00000171454
|
sphk2
|
sphingosine kinase 2 |
| chr3_+_26223376 | 1.72 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr20_-_17259573 | 1.70 |
ENSDART00000166397
|
AL928808.1
|
|
| chr8_+_26226044 | 1.67 |
ENSDART00000186503
|
wdr6
|
WD repeat domain 6 |
| chr7_-_32599669 | 1.67 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr14_-_48765262 | 1.64 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr19_+_9533008 | 1.62 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr6_-_37749711 | 1.61 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr5_+_28973264 | 1.59 |
ENSDART00000005638
|
stxbp1b
|
syntaxin binding protein 1b |
| chr13_-_15793585 | 1.59 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2 associated athanogene 5 |
| chr6_-_19341184 | 1.57 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr22_+_35516440 | 1.56 |
ENSDART00000184191
|
hykk.2
|
hydroxylysine kinase, tandem duplicate 2 |
| chr6_-_18698669 | 1.55 |
ENSDART00000168554
ENSDART00000175205 |
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr18_-_34156765 | 1.52 |
ENSDART00000059400
|
c18h3orf33
|
c18h3orf33 homolog (H. sapiens) |
| chr16_+_54210554 | 1.52 |
ENSDART00000172622
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr17_+_5793248 | 1.47 |
ENSDART00000153743
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr12_+_28854410 | 1.44 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr19_-_20446756 | 1.43 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr1_-_18336878 | 1.41 |
ENSDART00000054674
|
mtnr1aa
|
melatonin receptor 1A a |
| chr5_+_28972935 | 1.40 |
ENSDART00000193274
|
stxbp1b
|
syntaxin binding protein 1b |
| chr22_-_11606078 | 1.39 |
ENSDART00000146026
|
pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr19_+_34742706 | 1.38 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr2_+_35854242 | 1.37 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr3_+_27786601 | 1.36 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr15_-_915635 | 1.33 |
ENSDART00000179821
|
si:dkey-7i4.21
|
si:dkey-7i4.21 |
| chr4_-_16706776 | 1.32 |
ENSDART00000079461
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr6_-_22146258 | 1.31 |
ENSDART00000181700
|
gga1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr8_+_68864 | 1.30 |
ENSDART00000164574
|
prr16
|
proline rich 16 |
| chr1_-_13968153 | 1.30 |
ENSDART00000103383
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr4_+_14343706 | 1.29 |
ENSDART00000142845
|
prl2
|
prolactin 2 |
| chr1_+_44941031 | 1.29 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr23_+_38940700 | 1.28 |
ENSDART00000065331
|
sall4
|
spalt-like transcription factor 4 |
| chr4_-_3023182 | 1.28 |
ENSDART00000127894
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_+_27898091 | 1.28 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr11_-_23697217 | 1.27 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr11_-_41130239 | 1.26 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr14_+_31496543 | 1.24 |
ENSDART00000170683
|
phf6
|
PHD finger protein 6 |
| chr17_+_43468732 | 1.21 |
ENSDART00000055487
|
chmp3
|
charged multivesicular body protein 3 |
| chr4_+_25912654 | 1.19 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr22_+_10440991 | 1.19 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr6_-_13200585 | 1.15 |
ENSDART00000185321
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr16_+_10413551 | 1.12 |
ENSDART00000032877
ENSDART00000172827 |
ino80e
|
INO80 complex subunit E |
| chr5_+_30998764 | 1.10 |
ENSDART00000185783
ENSDART00000147874 |
ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr12_+_2870671 | 1.07 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr13_+_32636197 | 1.04 |
ENSDART00000141057
|
si:ch211-206k20.5
|
si:ch211-206k20.5 |
| chr3_+_17951790 | 1.03 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr4_+_76404572 | 1.03 |
ENSDART00000174020
|
FP074874.2
|
|
| chr14_+_12142828 | 1.02 |
ENSDART00000012438
|
hspa9
|
heat shock protein 9 |
| chr10_-_33379850 | 0.98 |
ENSDART00000186924
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr17_-_9995667 | 0.96 |
ENSDART00000148463
|
snx6
|
sorting nexin 6 |
| chr3_+_38793499 | 0.95 |
ENSDART00000083394
|
kcnj19a
|
potassium voltage-gated channel subfamily J member 19a |
| chr6_-_18249760 | 0.93 |
ENSDART00000192037
|
anapc11
|
APC11 anaphase promoting complex subunit 11 homolog (yeast) |
| chr25_-_21609645 | 0.91 |
ENSDART00000087817
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr1_+_8304904 | 0.88 |
ENSDART00000168631
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr5_+_41496490 | 0.87 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr12_-_9516981 | 0.87 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr23_-_27442544 | 0.86 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr4_-_17263210 | 0.86 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr5_-_18897482 | 0.85 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr13_-_26799244 | 0.82 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr3_+_57997980 | 0.80 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr5_+_20257225 | 0.78 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr22_-_8725768 | 0.75 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr21_-_37435162 | 0.74 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr5_+_69868911 | 0.74 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr6_-_32314913 | 0.70 |
ENSDART00000051779
|
atg4c
|
autophagy related 4C, cysteine peptidase |
| chr25_-_35996141 | 0.63 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr15_-_24178893 | 0.63 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr2_+_31806602 | 0.60 |
ENSDART00000086608
|
ranbp9
|
RAN binding protein 9 |
| chr21_+_45223194 | 0.60 |
ENSDART00000150902
|
si:ch73-269m14.3
|
si:ch73-269m14.3 |
| chr25_-_34845302 | 0.59 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr8_+_10823069 | 0.58 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr7_-_24644893 | 0.52 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr19_-_7441948 | 0.52 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr7_-_17779644 | 0.52 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr7_+_24153070 | 0.48 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr21_+_22423286 | 0.47 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr2_+_31308587 | 0.46 |
ENSDART00000027090
|
clul1
|
clusterin-like 1 (retinal) |
| chr3_-_13437799 | 0.46 |
ENSDART00000126455
ENSDART00000074068 |
dhps
|
deoxyhypusine synthase |
| chr6_+_14980761 | 0.45 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr5_+_33281105 | 0.44 |
ENSDART00000011337
|
surf1
|
surfeit 1 |
| chr18_-_14691727 | 0.43 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr7_-_5711197 | 0.43 |
ENSDART00000134367
|
si:dkey-10h3.2
|
si:dkey-10h3.2 |
| chr22_-_10440688 | 0.40 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr8_+_23009662 | 0.39 |
ENSDART00000030885
|
uckl1a
|
uridine-cytidine kinase 1-like 1a |
| chr21_-_29119560 | 0.36 |
ENSDART00000147367
|
havcr2
|
hepatitis A virus cellular receptor 2 |
| chr6_-_39218609 | 0.36 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
Network of associatons between targets according to the STRING database.
First level regulatory network of irx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.8 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 1.0 | 5.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 1.0 | 2.9 | GO:0042940 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.9 | 2.6 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.8 | 5.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.8 | 2.4 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.7 | 3.7 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.7 | 2.9 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.6 | 8.5 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.6 | 3.6 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.5 | 2.7 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.4 | 5.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.4 | 16.9 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.4 | 4.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.4 | 2.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.4 | 2.9 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.4 | 3.0 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.4 | 2.9 | GO:0036372 | opsin transport(GO:0036372) |
| 0.4 | 2.5 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 2.8 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.3 | 5.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.3 | 5.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 10.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 1.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 2.0 | GO:0042311 | vasodilation(GO:0042311) |
| 0.3 | 0.9 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.3 | 7.3 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.2 | 1.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 2.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 1.8 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 2.0 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.2 | 1.7 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 1.2 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 12.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 2.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 2.6 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.2 | 4.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 4.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 5.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 0.6 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.2 | 1.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 3.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 2.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 4.7 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 2.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 3.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 1.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 4.2 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.1 | 2.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.9 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 1.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.0 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 2.0 | GO:0039022 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.1 | 1.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 2.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.6 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 1.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 3.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.9 | GO:0051984 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 3.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.4 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.0 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 | 1.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 3.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 8.5 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.3 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 3.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 5.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.2 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.9 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.0 | 3.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.4 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 4.3 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 3.5 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 4.0 | GO:0050804 | modulation of synaptic transmission(GO:0050804) |
| 0.0 | 1.7 | GO:0010469 | regulation of receptor activity(GO:0010469) |
| 0.0 | 0.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 1.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0002031 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 2.6 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 1.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.7 | 2.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.6 | 2.9 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.4 | 5.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.4 | 2.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 2.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.3 | 4.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.3 | 2.7 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 2.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 2.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 2.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 19.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 3.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 3.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.2 | 4.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 3.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 2.8 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 10.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 10.8 | GO:0031968 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.1 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 2.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 3.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 5.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 5.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.7 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.1 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 1.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 4.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 4.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 9.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 4.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.0 | 2.9 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.8 | 5.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.7 | 2.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.7 | 2.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.6 | 1.7 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.6 | 8.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.5 | 2.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.5 | 2.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 12.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.4 | 3.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 3.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.4 | 2.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.4 | 1.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 2.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 2.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 10.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.6 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.3 | 4.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 3.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 5.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 4.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 3.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.2 | 2.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.2 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.7 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.1 | 2.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 2.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 3.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 1.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 6.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 15.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 6.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 2.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 11.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 4.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 5.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 32.3 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 5.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 5.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 3.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.2 | 4.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 3.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 2.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 3.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |