Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
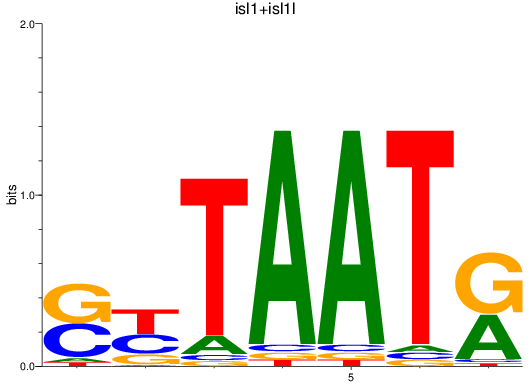
Results for isl1+isl1l
Z-value: 0.69
Transcription factors associated with isl1+isl1l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl1
|
ENSDARG00000004023 | ISL LIM homeobox 1 |
|
isl1l
|
ENSDARG00000021055 | islet1, like |
|
isl1l
|
ENSDARG00000110204 | islet1, like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl1 | dr11_v1_chr5_-_40734045_40734045 | 0.11 | 2.7e-01 | Click! |
| isl1l | dr11_v1_chr10_+_8680730_8680730 | -0.08 | 4.3e-01 | Click! |
Activity profile of isl1+isl1l motif
Sorted Z-values of isl1+isl1l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_11457500 | 6.63 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_-_30615901 | 4.97 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr10_-_2943474 | 4.04 |
ENSDART00000188698
|
oclna
|
occludin a |
| chr15_-_12011202 | 3.97 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr5_-_41831646 | 3.81 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr1_+_41588170 | 3.69 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr1_-_59348118 | 3.61 |
ENSDART00000170901
|
cyp3a65
|
cytochrome P450, family 3, subfamily A, polypeptide 65 |
| chr8_+_53064920 | 3.60 |
ENSDART00000164823
|
nadka
|
NAD kinase a |
| chr14_+_21222287 | 3.57 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr17_+_42274825 | 3.42 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr4_+_7508316 | 3.36 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr22_+_19220459 | 3.35 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr17_+_33375469 | 3.18 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr13_+_12456412 | 3.11 |
ENSDART00000057762
|
dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr7_+_13988075 | 3.07 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr1_-_25679339 | 2.99 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr4_+_76705830 | 2.93 |
ENSDART00000064312
|
ms4a17a.7
|
membrane-spanning 4-domains, subfamily A, member 17A.7 |
| chr24_-_32665283 | 2.93 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr14_-_36799280 | 2.61 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr19_+_10331325 | 2.56 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr10_+_35352435 | 2.53 |
ENSDART00000123936
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr17_+_39741926 | 2.37 |
ENSDART00000154996
ENSDART00000154599 |
si:dkey-229e3.2
|
si:dkey-229e3.2 |
| chr13_-_36391496 | 2.37 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr1_-_45146834 | 2.35 |
ENSDART00000144997
|
si:ch211-239f4.6
|
si:ch211-239f4.6 |
| chr3_+_15505275 | 2.31 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr23_+_26142807 | 2.25 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr17_-_2036850 | 2.18 |
ENSDART00000186048
ENSDART00000188838 ENSDART00000186423 |
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr10_+_35358675 | 2.17 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr13_+_46941930 | 2.16 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr23_+_26142613 | 2.14 |
ENSDART00000165046
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr5_-_41838354 | 2.06 |
ENSDART00000146793
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr17_+_6563307 | 1.91 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr12_-_36521767 | 1.89 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr4_-_75707991 | 1.87 |
ENSDART00000166358
ENSDART00000160021 |
si:dkey-165o8.2
|
si:dkey-165o8.2 |
| chr9_-_41507712 | 1.82 |
ENSDART00000135821
|
mfsd6b
|
major facilitator superfamily domain containing 6b |
| chr22_-_31020690 | 1.76 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr22_-_10121880 | 1.75 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr8_+_18830759 | 1.72 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr6_-_8580857 | 1.69 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr14_+_20893065 | 1.65 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr22_-_36875264 | 1.65 |
ENSDART00000137548
|
kng1
|
kininogen 1 |
| chr18_-_35459996 | 1.63 |
ENSDART00000141023
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr7_-_34192834 | 1.62 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr15_+_32642878 | 1.58 |
ENSDART00000159932
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr23_+_20431388 | 1.58 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr3_+_22377312 | 1.54 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr16_-_46619967 | 1.53 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr12_-_44307963 | 1.49 |
ENSDART00000161009
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr7_+_19482084 | 1.49 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr12_-_13205854 | 1.46 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr22_-_17653143 | 1.46 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr20_+_34770197 | 1.44 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr2_+_36112273 | 1.44 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr19_-_6193448 | 1.43 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr16_-_22192006 | 1.41 |
ENSDART00000163338
|
il6r
|
interleukin 6 receptor |
| chr3_-_32956808 | 1.41 |
ENSDART00000183902
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr10_-_36808348 | 1.40 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr5_+_32345187 | 1.39 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr8_-_14121634 | 1.38 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr4_+_2230701 | 1.38 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr15_-_25518084 | 1.37 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr4_+_9478500 | 1.35 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr8_+_21254192 | 1.29 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr7_-_54679595 | 1.27 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr23_-_45398622 | 1.27 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr6_-_27123327 | 1.25 |
ENSDART00000073881
|
agxta
|
alanine-glyoxylate aminotransferase a |
| chr15_-_44052927 | 1.24 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr5_-_64203101 | 1.24 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr10_+_38708099 | 1.24 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr6_-_53326421 | 1.21 |
ENSDART00000191740
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr13_+_15701849 | 1.21 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr22_-_3299100 | 1.20 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr22_-_621888 | 1.19 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr20_-_35012093 | 1.18 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr10_+_32104305 | 1.18 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr6_+_22326624 | 1.17 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr13_-_36582341 | 1.15 |
ENSDART00000137335
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr1_-_43727012 | 1.15 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr3_-_56871984 | 1.14 |
ENSDART00000189908
ENSDART00000183146 ENSDART00000183087 |
zgc:112148
|
zgc:112148 |
| chr14_-_30876299 | 1.12 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr5_+_22393501 | 1.11 |
ENSDART00000185276
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr12_-_19028046 | 1.11 |
ENSDART00000142253
ENSDART00000078484 |
rangap1b
|
Ran GTPase activating protein 1b |
| chr23_+_36653376 | 1.10 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr12_+_10062953 | 1.10 |
ENSDART00000148689
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr1_+_1915967 | 1.09 |
ENSDART00000131463
|
si:ch211-132g1.1
|
si:ch211-132g1.1 |
| chr25_-_10769039 | 1.08 |
ENSDART00000186758
ENSDART00000121724 |
lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr11_+_705727 | 1.08 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr20_+_23440632 | 1.08 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr2_-_50719170 | 1.08 |
ENSDART00000074943
|
xcr1b.1
|
chemokine (C motif) receptor 1b, duplicate 1 |
| chr20_-_22464250 | 1.07 |
ENSDART00000165904
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr4_-_890220 | 1.07 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr1_+_59328030 | 1.07 |
ENSDART00000172464
|
CABZ01052576.1
|
|
| chr19_-_47570672 | 1.06 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr8_-_30338872 | 1.05 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr25_+_30131055 | 1.04 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr22_+_19290199 | 1.04 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr14_-_32631013 | 1.04 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr20_+_2039518 | 1.03 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr15_+_11814969 | 1.03 |
ENSDART00000127248
|
FO704748.1
|
|
| chr5_+_31168096 | 1.03 |
ENSDART00000086443
ENSDART00000192271 |
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr13_-_337318 | 1.02 |
ENSDART00000166175
|
zgc:171534
|
zgc:171534 |
| chr16_+_52999778 | 1.02 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr19_+_8612839 | 1.00 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr16_-_29387215 | 0.99 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr20_-_45709990 | 0.99 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr6_-_40581376 | 0.99 |
ENSDART00000185412
|
tspo
|
translocator protein |
| chr5_+_25762271 | 0.96 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr16_-_30903930 | 0.96 |
ENSDART00000143996
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr17_+_47090497 | 0.95 |
ENSDART00000169038
ENSDART00000159292 |
zgc:103755
|
zgc:103755 |
| chr14_-_31856819 | 0.95 |
ENSDART00000003345
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr14_-_36437249 | 0.94 |
ENSDART00000016728
|
aga
|
aspartylglucosaminidase |
| chr14_+_16287968 | 0.94 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr3_-_8012847 | 0.93 |
ENSDART00000169673
|
si:ch211-175l6.1
|
si:ch211-175l6.1 |
| chr21_+_31253048 | 0.93 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr22_-_28644517 | 0.93 |
ENSDART00000189538
|
jagn1b
|
jagunal homolog 1b |
| chr9_+_54695981 | 0.93 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr4_+_71086017 | 0.92 |
ENSDART00000159113
|
si:dkeyp-80d11.10
|
si:dkeyp-80d11.10 |
| chr12_+_27034760 | 0.92 |
ENSDART00000181170
ENSDART00000153054 |
fbrs
|
fibrosin |
| chr24_-_33308045 | 0.92 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr3_-_8765165 | 0.91 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr1_-_9486214 | 0.90 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr21_+_26522571 | 0.90 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr10_-_10018120 | 0.90 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr22_+_6149121 | 0.89 |
ENSDART00000134067
|
si:dkey-19a16.5
|
si:dkey-19a16.5 |
| chr3_-_36209936 | 0.89 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr17_+_3124129 | 0.88 |
ENSDART00000155323
|
zgc:136872
|
zgc:136872 |
| chr4_-_12930086 | 0.88 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr9_+_8942258 | 0.87 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr11_-_438294 | 0.87 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr2_+_20604775 | 0.87 |
ENSDART00000131501
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr8_+_20140321 | 0.86 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr15_-_43625549 | 0.86 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr15_-_39969988 | 0.86 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr13_-_42536642 | 0.85 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr23_+_20431140 | 0.85 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr16_-_55259199 | 0.85 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr12_+_27034594 | 0.84 |
ENSDART00000111679
|
fbrs
|
fibrosin |
| chr20_-_40360571 | 0.82 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr11_+_27347076 | 0.82 |
ENSDART00000173383
|
fbln2
|
fibulin 2 |
| chr13_+_33689740 | 0.81 |
ENSDART00000161904
|
ephx5
|
epoxide hydrolase 5 |
| chr9_-_14137295 | 0.81 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr23_-_31372639 | 0.78 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr14_+_23709134 | 0.76 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr16_+_53203370 | 0.75 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr12_+_22672323 | 0.73 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr6_-_436658 | 0.73 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr9_+_3283608 | 0.73 |
ENSDART00000192275
|
hat1
|
histone acetyltransferase 1 |
| chr1_+_9994811 | 0.72 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr19_-_7272921 | 0.72 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr12_-_13205572 | 0.72 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr18_+_7612438 | 0.72 |
ENSDART00000175288
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr9_-_35633827 | 0.72 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr7_+_38897836 | 0.72 |
ENSDART00000024330
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr5_-_19006290 | 0.69 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr23_-_38497705 | 0.69 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr15_-_15983428 | 0.69 |
ENSDART00000115129
|
synrg
|
synergin, gamma |
| chr22_-_16755885 | 0.69 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr12_+_28910762 | 0.68 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr5_-_33281046 | 0.68 |
ENSDART00000051344
ENSDART00000138116 |
surf6
|
surfeit 6 |
| chr24_+_36636208 | 0.67 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr7_+_3872189 | 0.66 |
ENSDART00000064221
|
si:dkey-88n24.5
|
si:dkey-88n24.5 |
| chr18_+_48953963 | 0.66 |
ENSDART00000158104
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr11_-_1400507 | 0.62 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr21_+_2506013 | 0.61 |
ENSDART00000162351
|
hmgcrb
|
3-hydroxy-3-methylglutaryl-CoA reductase b |
| chr4_-_14192254 | 0.59 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr7_+_26998169 | 0.59 |
ENSDART00000128110
ENSDART00000101018 |
caprin1a
|
cell cycle associated protein 1a |
| chr20_+_29209926 | 0.59 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr14_+_1240419 | 0.59 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr20_-_45772306 | 0.58 |
ENSDART00000062092
|
trmt6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr3_+_8224622 | 0.58 |
ENSDART00000138670
|
si:ch73-379f5.5
|
si:ch73-379f5.5 |
| chr5_-_36597612 | 0.57 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr20_-_25522911 | 0.57 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr6_+_15268685 | 0.57 |
ENSDART00000128090
ENSDART00000154417 |
ecrg4b
|
esophageal cancer related gene 4b |
| chr21_-_30408775 | 0.57 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr5_-_13076779 | 0.57 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr2_+_10007113 | 0.55 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr13_-_11971148 | 0.55 |
ENSDART00000066230
ENSDART00000185614 |
ARL3 (1 of many)
|
zgc:110197 |
| chr18_+_20677090 | 0.54 |
ENSDART00000060243
|
cftr
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr15_-_1962326 | 0.54 |
ENSDART00000179669
|
dock10
|
dedicator of cytokinesis 10 |
| chr20_-_5400395 | 0.53 |
ENSDART00000168103
|
snw1
|
SNW domain containing 1 |
| chr3_-_23513155 | 0.53 |
ENSDART00000170200
|
BX682558.1
|
|
| chr22_-_26852516 | 0.53 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr14_-_1454045 | 0.52 |
ENSDART00000161460
|
pmt
|
phosphoethanolamine methyltransferase |
| chr3_-_18737126 | 0.52 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr11_-_37997419 | 0.52 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr25_+_32390794 | 0.52 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr15_+_1199407 | 0.52 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr2_-_10821053 | 0.52 |
ENSDART00000056034
|
rpap2
|
RNA polymerase II associated protein 2 |
| chr5_+_58687541 | 0.52 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr5_+_66250856 | 0.51 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr1_-_25144439 | 0.51 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr3_-_11008532 | 0.51 |
ENSDART00000165086
|
CR382337.3
|
|
| chr16_+_9400661 | 0.50 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr11_-_18107447 | 0.50 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr4_-_15103646 | 0.50 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr4_-_71214516 | 0.50 |
ENSDART00000169997
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr16_-_36979592 | 0.48 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr9_+_30478768 | 0.48 |
ENSDART00000101097
|
acp6
|
acid phosphatase 6, lysophosphatidic |
| chr18_-_50839033 | 0.47 |
ENSDART00000169773
|
ddb1
|
damage-specific DNA binding protein 1 |
| chr16_+_9762261 | 0.47 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl1+isl1l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.6 | 1.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.5 | 2.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.5 | 2.2 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.5 | 3.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.5 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 1.8 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.4 | 1.2 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.3 | 0.9 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.3 | 0.8 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.3 | 0.8 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 1.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 1.5 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 | 1.7 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.2 | 1.6 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.2 | 0.9 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 | 0.9 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 1.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 | 3.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 1.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.2 | 1.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.2 | 1.4 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 3.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 1.2 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.2 | 0.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 0.5 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.2 | 1.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 0.5 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.2 | 1.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.2 | 0.5 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 1.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 2.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 1.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.9 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.9 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 2.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.3 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 1.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 3.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 2.9 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.3 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 1.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 2.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.8 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 1.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.5 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 4.2 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 1.0 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.8 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.6 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 4.3 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 4.5 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.0 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 1.1 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.2 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 1.2 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.1 | GO:2000047 | cell-cell adhesion mediated by cadherin(GO:0044331) regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.3 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 1.4 | GO:0044218 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.4 | 1.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 1.8 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 0.9 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 3.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 4.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.1 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.7 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 6.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0022627 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.6 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.5 | 1.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.4 | 1.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 1.8 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.3 | 1.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 3.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 1.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 0.8 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.3 | 1.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 0.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 1.4 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 1.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.1 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.7 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 3.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.0 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 3.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 2.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 0.7 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.9 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.3 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 4.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 1.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 1.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 8.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.7 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.2 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 4.0 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.3 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.9 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 4.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 2.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 3.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 3.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.0 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME GLOBAL GENOMIC NER GG NER | Genes involved in Global Genomic NER (GG-NER) |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |