Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
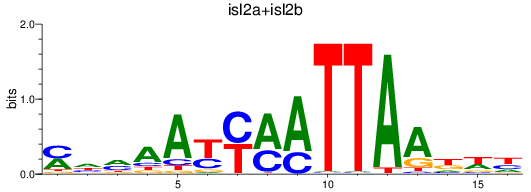
Results for isl2a+isl2b
Z-value: 0.59
Transcription factors associated with isl2a+isl2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl2a
|
ENSDARG00000003971 | ISL LIM homeobox 2a |
|
isl2b
|
ENSDARG00000053499 | ISL LIM homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl2b | dr11_v1_chr7_+_29992889_29992889 | 0.22 | 3.2e-02 | Click! |
| isl2a | dr11_v1_chr25_-_32751982_32751982 | 0.09 | 4.0e-01 | Click! |
Activity profile of isl2a+isl2b motif
Sorted Z-values of isl2a+isl2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45094599 | 4.28 |
ENSDART00000155479
|
si:rp71-77l1.1
|
si:rp71-77l1.1 |
| chr18_+_30847237 | 4.14 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr25_+_21833287 | 3.78 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr15_+_46356879 | 3.76 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr20_-_5291012 | 3.50 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr15_+_46357080 | 3.49 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr15_-_2640966 | 3.42 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr12_+_17154655 | 3.28 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr19_+_1688727 | 2.83 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr24_-_26310854 | 2.62 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr20_+_41021054 | 2.45 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr21_+_25236297 | 2.43 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr23_-_33350990 | 2.41 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr15_-_46779934 | 2.33 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr5_+_58550291 | 2.15 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr8_+_52637507 | 2.13 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr25_+_3994823 | 2.06 |
ENSDART00000154020
|
eps8l2
|
EPS8 like 2 |
| chr21_-_44081540 | 2.02 |
ENSDART00000130833
|
FO704810.1
|
|
| chr17_+_15433518 | 1.98 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr22_-_16154771 | 1.90 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr22_+_19552987 | 1.87 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr1_+_52792439 | 1.75 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr6_-_35046735 | 1.75 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr18_-_48547564 | 1.72 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr17_+_15433671 | 1.72 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr23_-_31645760 | 1.71 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr18_-_15932704 | 1.69 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr21_+_11385031 | 1.64 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr8_-_50147948 | 1.63 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr14_-_4145594 | 1.61 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr13_-_36391496 | 1.52 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr7_+_66822229 | 1.51 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr16_+_54209504 | 1.47 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr3_-_25268751 | 1.45 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr7_+_22657566 | 1.45 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr5_-_19861766 | 1.44 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr11_+_24002503 | 1.42 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr19_-_5769728 | 1.41 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr11_+_24001993 | 1.39 |
ENSDART00000168215
|
chia.2
|
chitinase, acidic.2 |
| chr13_-_33639050 | 1.37 |
ENSDART00000133073
|
rrbp1a
|
ribosome binding protein 1a |
| chr10_+_29850330 | 1.34 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr3_+_26342768 | 1.25 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr10_-_45210184 | 1.22 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr21_-_35419486 | 1.22 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr12_-_4532066 | 1.21 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr5_+_69785990 | 1.14 |
ENSDART00000162057
ENSDART00000166893 |
kmt5ab
|
lysine methyltransferase 5Ab |
| chr19_+_16015881 | 1.14 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr2_+_36112273 | 1.13 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr5_+_60590796 | 1.13 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr22_-_5252005 | 1.12 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr6_-_12588044 | 1.11 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr16_-_39477746 | 1.10 |
ENSDART00000102525
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr6_-_16717878 | 1.10 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr1_-_9641845 | 1.08 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr25_-_21031007 | 1.07 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr23_+_36460239 | 1.06 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr16_+_23303859 | 1.03 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr15_-_43284021 | 1.02 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr15_+_36115955 | 1.02 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr8_+_25913787 | 1.01 |
ENSDART00000190257
ENSDART00000062515 |
sema3h
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3H |
| chr2_+_2169337 | 1.00 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr4_-_17669881 | 0.96 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr24_+_12989727 | 0.94 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr12_+_18899396 | 0.91 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr5_+_69786215 | 0.88 |
ENSDART00000165956
|
kmt5ab
|
lysine methyltransferase 5Ab |
| chr5_+_56023186 | 0.87 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr16_-_17072440 | 0.87 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr12_+_16168342 | 0.85 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr15_+_25438714 | 0.84 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr23_-_36724575 | 0.83 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr22_-_10165446 | 0.83 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr1_-_55873178 | 0.82 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr2_-_38284648 | 0.82 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr21_+_27513859 | 0.81 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr6_+_39905021 | 0.80 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr12_+_22560067 | 0.80 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr22_-_36530902 | 0.78 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_+_3681116 | 0.78 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr3_+_28939759 | 0.78 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr3_+_14388010 | 0.77 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr21_+_45841731 | 0.76 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr14_+_21722235 | 0.76 |
ENSDART00000183667
|
stx3a
|
syntaxin 3A |
| chr9_+_24008879 | 0.73 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr4_-_77218637 | 0.72 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr7_-_48665305 | 0.71 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr24_-_6078222 | 0.71 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr12_+_10062953 | 0.71 |
ENSDART00000148689
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr13_-_30996072 | 0.70 |
ENSDART00000181661
|
wdfy4
|
WDFY family member 4 |
| chr3_-_19368435 | 0.70 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr25_+_35212919 | 0.70 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr19_+_7424347 | 0.65 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr21_+_7100442 | 0.63 |
ENSDART00000163869
|
ppp1r26
|
protein phosphatase 1, regulatory subunit 26 |
| chr13_+_7049823 | 0.61 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr6_-_34838397 | 0.59 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr18_+_17537344 | 0.59 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr24_+_19415124 | 0.59 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr20_+_25586099 | 0.58 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr18_+_13275735 | 0.57 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr13_+_646700 | 0.56 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr22_-_17482282 | 0.56 |
ENSDART00000105453
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr3_+_7808459 | 0.55 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr3_+_32553714 | 0.54 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr17_-_16422654 | 0.54 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr8_-_11324143 | 0.54 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr15_-_21702317 | 0.54 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr3_-_26787430 | 0.53 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr19_+_31904836 | 0.53 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr21_-_35534401 | 0.52 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr9_+_21793565 | 0.52 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr19_+_16016038 | 0.51 |
ENSDART00000131319
|
ctps1a
|
CTP synthase 1a |
| chr18_+_17537975 | 0.51 |
ENSDART00000179783
|
nup93
|
nucleoporin 93 |
| chr23_-_18024543 | 0.51 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr2_-_59205393 | 0.51 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr7_+_39166460 | 0.50 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr11_-_45138857 | 0.50 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr12_-_9516981 | 0.49 |
ENSDART00000106285
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr7_-_2047639 | 0.48 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr16_-_48400639 | 0.46 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr8_+_31717175 | 0.46 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr11_+_29856032 | 0.46 |
ENSDART00000079150
|
grpr
|
gastrin-releasing peptide receptor |
| chr2_+_10771787 | 0.46 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr24_-_12770357 | 0.44 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr10_-_34916208 | 0.44 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr7_+_24573721 | 0.43 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr6_-_8311044 | 0.43 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr21_-_10886709 | 0.40 |
ENSDART00000134408
|
si:dkey-277m11.2
|
si:dkey-277m11.2 |
| chr4_-_75158035 | 0.39 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr19_+_33093577 | 0.38 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr24_-_32173754 | 0.38 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr4_-_1801519 | 0.38 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr11_-_29737088 | 0.37 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr9_+_32069989 | 0.37 |
ENSDART00000139540
|
si:dkey-83m22.7
|
si:dkey-83m22.7 |
| chr4_-_46915962 | 0.34 |
ENSDART00000169555
|
si:ch211-134c10.1
|
si:ch211-134c10.1 |
| chr3_+_32532645 | 0.34 |
ENSDART00000055312
ENSDART00000150981 |
nosip
|
nitric oxide synthase interacting protein |
| chr15_+_1766734 | 0.34 |
ENSDART00000168250
|
cul3b
|
cullin 3b |
| chr4_-_49582758 | 0.34 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr10_+_44956660 | 0.34 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr1_+_55608520 | 0.33 |
ENSDART00000152307
|
adgre18
|
adhesion G protein-coupled receptor E18 |
| chr16_+_11660839 | 0.33 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr7_-_50410524 | 0.33 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr2_-_293793 | 0.33 |
ENSDART00000082091
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr16_-_28658341 | 0.33 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr21_-_37194839 | 0.32 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr1_+_55758257 | 0.32 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr6_+_40951227 | 0.28 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr6_+_12527725 | 0.28 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr3_+_32532000 | 0.28 |
ENSDART00000165909
|
nosip
|
nitric oxide synthase interacting protein |
| chr21_-_35325466 | 0.27 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr10_-_43771447 | 0.27 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr5_+_32076109 | 0.26 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr18_+_43891051 | 0.26 |
ENSDART00000024213
|
cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr3_-_54846444 | 0.25 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr13_+_15657911 | 0.24 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr1_-_669717 | 0.24 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr25_+_22320738 | 0.23 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr7_-_66868543 | 0.23 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr4_-_9909371 | 0.23 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr2_-_22927958 | 0.23 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr2_+_10878406 | 0.22 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr5_-_42083363 | 0.22 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr5_+_22791686 | 0.21 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr23_+_31981796 | 0.20 |
ENSDART00000187080
|
pan2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr21_+_43172506 | 0.17 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr2_-_293036 | 0.15 |
ENSDART00000171629
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr22_-_17482810 | 0.15 |
ENSDART00000171293
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr16_-_10223741 | 0.15 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr21_+_4256291 | 0.15 |
ENSDART00000148138
|
lrrc8aa
|
leucine rich repeat containing 8 VRAC subunit Aa |
| chr7_+_60079302 | 0.15 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr23_+_9353552 | 0.15 |
ENSDART00000163298
|
BX511246.1
|
|
| chr8_+_31716872 | 0.14 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr17_-_2834764 | 0.14 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr11_-_10456387 | 0.13 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr2_-_10877765 | 0.12 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr11_+_31324335 | 0.11 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr9_+_54039006 | 0.09 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr23_+_32406461 | 0.09 |
ENSDART00000179878
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr2_+_20605925 | 0.09 |
ENSDART00000191510
|
olfml2bb
|
olfactomedin-like 2Bb |
| chr17_-_4245902 | 0.09 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr7_-_7797654 | 0.09 |
ENSDART00000084503
ENSDART00000192779 ENSDART00000173079 |
trmt10b
|
tRNA methyltransferase 10B |
| chr24_-_3477103 | 0.08 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr21_+_11749097 | 0.08 |
ENSDART00000102408
ENSDART00000102404 |
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr2_-_1622641 | 0.07 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr1_-_50859053 | 0.07 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr13_-_42560662 | 0.07 |
ENSDART00000124898
|
CR792417.1
|
|
| chr16_-_21140097 | 0.05 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr11_-_10456553 | 0.04 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr23_-_3511630 | 0.03 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr17_-_29224908 | 0.00 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr19_+_24872159 | 0.00 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl2a+isl2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.2 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.5 | 2.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 1.7 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.4 | 1.2 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.4 | 2.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 2.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 3.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 3.8 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 0.9 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 1.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 1.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 2.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 0.9 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 1.0 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.8 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 1.6 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 1.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.5 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 1.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 2.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.0 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 1.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.6 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 1.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 2.6 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 0.4 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 2.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.6 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.8 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 3.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.9 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.1 | GO:0043687 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.1 | 0.6 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.8 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 1.2 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.5 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 3.6 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.5 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 3.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.2 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.3 | 1.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.6 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 0.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 1.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 3.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.5 | 1.6 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 5.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.4 | 1.7 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.4 | 1.8 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.3 | 1.2 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.3 | 3.8 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 3.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 2.5 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 0.6 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.2 | 0.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 0.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.2 | 1.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 2.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.3 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.8 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 1.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 2.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.9 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.0 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.2 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 2.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 4.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0042562 | hormone binding(GO:0042562) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 4.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 1.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |