Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
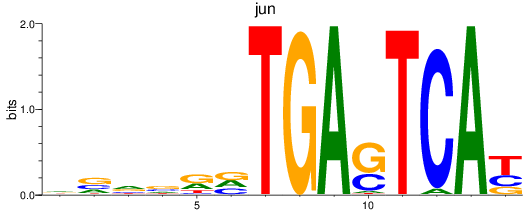
Results for jun
Z-value: 0.90
Transcription factors associated with jun
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jun
|
ENSDARG00000043531 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jun | dr11_v1_chr20_+_15552657_15552657 | 0.18 | 7.9e-02 | Click! |
Activity profile of jun motif
Sorted Z-values of jun motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_26880668 | 14.05 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr20_+_28364742 | 13.60 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr16_+_23403602 | 11.35 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr7_+_25033924 | 8.80 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr21_+_25765734 | 7.60 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr19_+_24488403 | 7.47 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr16_-_24832038 | 7.42 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr1_-_25679339 | 7.22 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr13_+_23988442 | 6.34 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr5_+_45677781 | 6.07 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr10_+_28428222 | 5.97 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr16_-_36834505 | 5.66 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr1_-_58913813 | 5.61 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
| chr1_+_59088205 | 5.56 |
ENSDART00000150649
ENSDART00000100197 |
zgc:173915
|
zgc:173915 |
| chr1_-_25177086 | 5.48 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr7_+_33172066 | 5.43 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr22_-_10110959 | 5.31 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr16_-_21785261 | 5.27 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr1_+_59090743 | 5.26 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr1_+_14253118 | 5.26 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr1_+_59090972 | 5.07 |
ENSDART00000171497
|
mfap4
|
microfibril associated protein 4 |
| chr16_+_46410520 | 4.99 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr8_-_38201415 | 4.94 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr5_-_30615901 | 4.90 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr25_+_10410620 | 4.84 |
ENSDART00000151886
|
ehf
|
ets homologous factor |
| chr21_+_33459524 | 4.82 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr4_+_25181572 | 4.66 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr19_+_7567763 | 4.53 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr1_+_59073436 | 4.49 |
ENSDART00000161642
|
MFAP4 (1 of many)
|
si:zfos-2330d3.3 |
| chr10_+_187760 | 4.46 |
ENSDART00000161007
ENSDART00000160651 |
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr4_+_16715267 | 4.39 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr2_+_15100742 | 4.33 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr8_-_18667693 | 4.30 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr15_+_963292 | 4.25 |
ENSDART00000156586
|
alox5b.2
|
arachidonate 5-lipoxygenase b, tandem duplicate 2 |
| chr3_+_32118670 | 4.21 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr12_+_25945560 | 4.21 |
ENSDART00000109799
|
mmrn2b
|
multimerin 2b |
| chr11_-_24681292 | 4.19 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr16_+_11558868 | 4.16 |
ENSDART00000112497
ENSDART00000180445 |
zgc:198329
|
zgc:198329 |
| chr6_-_49063085 | 4.14 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr6_-_40842768 | 4.13 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr3_-_26017592 | 4.08 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr15_-_46779934 | 4.08 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr14_+_33882973 | 4.01 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr23_+_17522867 | 3.89 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr17_-_53359028 | 3.78 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr2_+_30379650 | 3.76 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr4_-_20181964 | 3.72 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr3_-_26017831 | 3.68 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr7_+_14005111 | 3.63 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr14_+_30774032 | 3.58 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr14_+_30774515 | 3.56 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr10_+_22771176 | 3.52 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr2_-_898899 | 3.51 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr5_+_24089334 | 3.51 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr1_-_59077650 | 3.47 |
ENSDART00000043516
|
MFAP4 (1 of many)
|
si:zfos-2330d3.1 |
| chr16_-_45225520 | 3.47 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr25_+_18583877 | 3.46 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr2_+_25839940 | 3.32 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr19_-_5332784 | 3.30 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr22_-_24297510 | 3.30 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr3_+_21189766 | 3.27 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr25_+_29474583 | 3.27 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr5_+_37087583 | 3.25 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr7_+_69019851 | 3.24 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr8_+_6576940 | 3.21 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr7_+_34305903 | 3.20 |
ENSDART00000173575
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr3_+_22375596 | 3.18 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr3_-_15734530 | 3.17 |
ENSDART00000141142
|
mvp
|
major vault protein |
| chr7_-_28696556 | 3.09 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr8_+_5024468 | 3.09 |
ENSDART00000030938
|
adra1aa
|
adrenoceptor alpha 1Aa |
| chr7_-_29534001 | 3.08 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr3_-_15734358 | 3.07 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr25_-_22191733 | 3.05 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr22_+_16497670 | 3.04 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr7_+_22688781 | 3.03 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr10_+_38610741 | 2.94 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr23_+_25856541 | 2.90 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr25_+_32474031 | 2.88 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr21_+_26612777 | 2.86 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr19_-_7420867 | 2.86 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr8_-_44463985 | 2.84 |
ENSDART00000016845
|
mhc1lba
|
major histocompatibility complex class I LBA |
| chr20_-_34670236 | 2.83 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr20_-_35578435 | 2.78 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr10_+_26747755 | 2.76 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr3_+_3598555 | 2.75 |
ENSDART00000191152
|
CR589947.3
|
|
| chr1_-_55116453 | 2.74 |
ENSDART00000142348
|
sertad2a
|
SERTA domain containing 2a |
| chr10_-_3295197 | 2.73 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr3_+_46762703 | 2.72 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr23_-_1017605 | 2.72 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr13_+_829585 | 2.71 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr21_-_2415808 | 2.71 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr22_+_4442473 | 2.70 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr22_-_24858042 | 2.70 |
ENSDART00000137998
ENSDART00000078216 ENSDART00000138378 |
vtg7
|
vitellogenin 7 |
| chr14_+_30774894 | 2.69 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr8_-_11170114 | 2.67 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr20_+_9683994 | 2.65 |
ENSDART00000053831
|
ptger2b
|
prostaglandin E receptor 2b (subtype EP2) |
| chr7_-_32981559 | 2.64 |
ENSDART00000175614
|
pkp3b
|
plakophilin 3b |
| chr23_+_4324625 | 2.63 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr5_-_25582721 | 2.62 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr16_+_23398369 | 2.59 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr5_-_20123002 | 2.58 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr3_-_58189429 | 2.57 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr11_-_23501467 | 2.52 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr6_+_42475730 | 2.50 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr13_-_37122217 | 2.49 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr14_-_1538600 | 2.46 |
ENSDART00000180925
|
CABZ01109480.1
|
|
| chr5_-_26879302 | 2.45 |
ENSDART00000098571
ENSDART00000139086 |
zgc:64051
|
zgc:64051 |
| chr3_-_48259289 | 2.41 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr15_+_3766101 | 2.41 |
ENSDART00000042580
ENSDART00000112698 ENSDART00000187035 ENSDART00000165571 ENSDART00000121752 |
RNF14 (1 of many)
|
zmp:0000000524 |
| chr22_-_606067 | 2.39 |
ENSDART00000136722
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr4_-_17822447 | 2.39 |
ENSDART00000185502
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr13_-_37649595 | 2.39 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr2_-_37862380 | 2.38 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr24_-_33366188 | 2.36 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr8_-_30204650 | 2.33 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr22_-_23006842 | 2.33 |
ENSDART00000105607
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr15_-_28223757 | 2.28 |
ENSDART00000110969
ENSDART00000138401 |
scarf1
|
scavenger receptor class F, member 1 |
| chr24_+_24726956 | 2.27 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr8_+_8671229 | 2.22 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr12_+_20587179 | 2.21 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr20_+_25625872 | 2.19 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr23_-_1017428 | 2.19 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr8_-_23573084 | 2.19 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr24_-_23839647 | 2.18 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr20_+_7084154 | 2.17 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr22_+_19405517 | 2.16 |
ENSDART00000138245
ENSDART00000155144 |
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr19_+_37120491 | 2.15 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr2_+_36109002 | 2.13 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr1_-_57501299 | 2.13 |
ENSDART00000080600
|
zgc:171470
|
zgc:171470 |
| chr20_+_6543674 | 2.11 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr23_+_28377360 | 2.08 |
ENSDART00000014983
ENSDART00000128831 ENSDART00000135178 ENSDART00000138621 |
zgc:153867
|
zgc:153867 |
| chr2_+_25839650 | 2.07 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr25_+_29474982 | 2.05 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr19_+_348729 | 2.03 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr10_-_22095505 | 2.02 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr15_-_38129845 | 2.02 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr2_+_25378457 | 2.02 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr5_+_15819651 | 2.01 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr21_+_28747069 | 2.00 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr23_-_45398622 | 2.00 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr4_+_7822773 | 1.99 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr2_+_25840463 | 1.99 |
ENSDART00000125178
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr5_+_50953240 | 1.95 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr16_+_49005321 | 1.95 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr24_-_30275204 | 1.93 |
ENSDART00000164187
|
snx7
|
sorting nexin 7 |
| chr6_+_56141852 | 1.93 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr23_-_36316352 | 1.93 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr5_+_42141917 | 1.90 |
ENSDART00000172201
ENSDART00000140743 |
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr1_+_6640437 | 1.89 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr4_-_6809323 | 1.88 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr8_-_38403018 | 1.88 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr19_+_43753995 | 1.88 |
ENSDART00000058504
|
SLC17A3
|
si:ch1073-513e17.1 |
| chr8_-_38105053 | 1.86 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr11_+_13630107 | 1.85 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr23_-_5783421 | 1.84 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr19_+_24575077 | 1.83 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr1_-_34335752 | 1.81 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr10_-_39283883 | 1.79 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr5_-_1047504 | 1.79 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr10_+_38593645 | 1.77 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr13_-_51846224 | 1.74 |
ENSDART00000184663
|
LT631684.2
|
|
| chr8_-_2616326 | 1.73 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr6_-_35439406 | 1.71 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr13_-_15982707 | 1.71 |
ENSDART00000186911
ENSDART00000181072 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr6_-_43028896 | 1.71 |
ENSDART00000149977
|
glyctk
|
glycerate kinase |
| chr4_+_77971104 | 1.70 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr8_+_32406885 | 1.69 |
ENSDART00000167600
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr11_-_18253111 | 1.68 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr5_-_57480660 | 1.68 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr3_-_39648021 | 1.68 |
ENSDART00000055171
|
grapa
|
GRB2-related adaptor protein a |
| chr17_-_15600455 | 1.67 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr16_+_37876779 | 1.66 |
ENSDART00000140148
|
si:ch211-198c19.1
|
si:ch211-198c19.1 |
| chr3_+_1179601 | 1.65 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr20_+_51479263 | 1.65 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr19_+_2670130 | 1.62 |
ENSDART00000152152
|
si:ch73-24k9.2
|
si:ch73-24k9.2 |
| chr19_-_10432134 | 1.60 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr4_+_77933084 | 1.56 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_-_34616599 | 1.56 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr6_-_59357256 | 1.56 |
ENSDART00000074534
|
fam210b
|
family with sequence similarity 210, member B |
| chr1_-_59567685 | 1.55 |
ENSDART00000159144
ENSDART00000182913 |
zmp:0000001082
|
zmp:0000001082 |
| chr3_-_34034498 | 1.55 |
ENSDART00000151145
|
ighv11-2
|
immunoglobulin heavy variable 11-2 |
| chr6_-_43449013 | 1.54 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr6_-_33980881 | 1.53 |
ENSDART00000057297
|
mpl
|
MPL proto-oncogene, thrombopoietin receptor |
| chr6_+_48348415 | 1.52 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr4_+_357810 | 1.51 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr13_+_35339182 | 1.50 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr25_+_30196039 | 1.50 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr11_+_13629528 | 1.48 |
ENSDART00000186509
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr1_+_54655160 | 1.48 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr23_+_19606291 | 1.47 |
ENSDART00000139415
|
flnb
|
filamin B, beta (actin binding protein 278) |
| chr10_-_43850304 | 1.46 |
ENSDART00000180052
|
tlr8b
|
toll-like receptor 8b |
| chr23_+_25232711 | 1.46 |
ENSDART00000128510
|
erbb3b
|
erb-b2 receptor tyrosine kinase 3b |
| chr18_+_8320165 | 1.45 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr2_-_37277626 | 1.45 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr13_-_16222388 | 1.44 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr21_+_28747236 | 1.42 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr24_-_23998897 | 1.41 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr21_-_21020708 | 1.40 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr7_+_34786591 | 1.38 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr6_-_37744430 | 1.38 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
Network of associatons between targets according to the STRING database.
First level regulatory network of jun
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 2.6 | 7.8 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 1.8 | 5.3 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 1.6 | 4.8 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 1.4 | 2.7 | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) myeloid leukocyte cytokine production(GO:0061082) |
| 1.3 | 21.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.2 | 3.5 | GO:0061317 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 1.2 | 3.5 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 1.2 | 5.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 1.1 | 7.4 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 1.0 | 2.9 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.7 | 2.7 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.7 | 5.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.7 | 2.6 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.6 | 4.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.6 | 1.9 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.6 | 3.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.5 | 2.7 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.5 | 3.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.5 | 7.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.5 | 1.9 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.5 | 3.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 3.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.4 | 2.6 | GO:0006925 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 | 1.3 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.4 | 6.3 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.4 | 2.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.4 | 4.2 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 6.0 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.3 | 3.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 2.3 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.3 | 2.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.3 | 6.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.3 | 3.6 | GO:0016486 | peptide hormone processing(GO:0016486) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.3 | 1.0 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 2.7 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.2 | 2.9 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.2 | 0.7 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.2 | 12.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.2 | 1.1 | GO:0032527 | retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.2 | 1.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 1.0 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 1.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 2.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 1.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 1.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 2.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.2 | 1.1 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.2 | 5.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 0.9 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 8.1 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.2 | 1.9 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.1 | 1.0 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 7.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 2.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.7 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 1.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.0 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 1.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 2.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.1 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.5 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 2.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 4.8 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.4 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 7.1 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.9 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.0 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.5 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 1.7 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.5 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 1.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 2.7 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 5.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.3 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 2.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.5 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 2.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 3.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 0.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.3 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 4.7 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.1 | 7.4 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 2.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.6 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 4.2 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 0.5 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.3 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.8 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 3.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.9 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 1.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.9 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 1.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.0 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 2.0 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.8 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.1 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.0 | 1.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 5.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.2 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.6 | GO:0007164 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.9 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 3.2 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 1.0 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 2.4 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.9 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 5.5 | GO:0009116 | nucleoside metabolic process(GO:0009116) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.6 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 1.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.5 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 2.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.6 | 4.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 1.9 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.4 | 1.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 7.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 4.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 5.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 1.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 1.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 0.9 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 1.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 2.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 7.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.5 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 5.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.7 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 3.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 4.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 11.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 7.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.5 | GO:0060293 | P granule(GO:0043186) germ plasm(GO:0060293) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 62.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.6 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 3.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 3.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 3.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 12.9 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.3 | 5.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 1.3 | 7.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.1 | 3.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.0 | 2.9 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.7 | 4.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.7 | 18.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.6 | 4.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.6 | 1.8 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.6 | 5.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.6 | 5.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.5 | 1.5 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.4 | 5.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 5.7 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.4 | 2.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 3.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 1.7 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.4 | 3.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.4 | 2.7 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.4 | 1.9 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.3 | 1.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 23.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 1.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.3 | 1.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.3 | 0.9 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 1.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 2.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.3 | 6.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 1.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 5.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.9 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 5.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 1.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 4.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 5.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 1.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 6.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.5 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 1.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 2.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 6.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 8.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.5 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 9.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 10.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.9 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 2.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 3.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.7 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 3.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 15.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.5 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 3.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 4.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 4.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.0 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 2.2 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 3.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 3.7 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 4.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.7 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 5.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 5.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 6.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 3.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 13.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 4.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 9.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 5.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 9.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.4 | 3.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 2.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 2.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 2.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 3.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.2 | 3.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 6.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.2 | 2.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 1.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 9.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 10.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.5 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |