Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
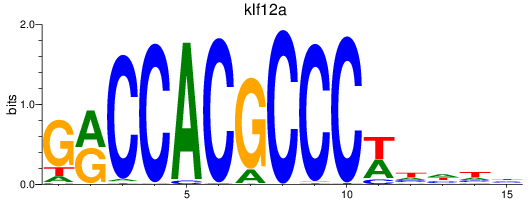
Results for klf12a
Z-value: 1.02
Transcription factors associated with klf12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12a
|
ENSDARG00000015312 | Kruppel-like factor 12a |
|
klf12a
|
ENSDARG00000115152 | Kruppel-like factor 12a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12a | dr11_v1_chr1_+_34527213_34527213 | 0.22 | 3.1e-02 | Click! |
Activity profile of klf12a motif
Sorted Z-values of klf12a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_58221163 | 20.32 |
ENSDART00000157939
|
FO704813.1
|
|
| chr1_-_59176949 | 19.02 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr10_+_15777064 | 16.06 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr10_+_1668106 | 15.16 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr10_+_19554604 | 14.04 |
ENSDART00000063806
|
atp6v1b2
|
ATPase H+ transporting V1 subunit B2 |
| chr23_+_44732863 | 13.73 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr5_-_46273938 | 13.67 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr16_-_6821927 | 13.35 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr9_+_42095220 | 13.26 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr5_+_37056818 | 13.23 |
ENSDART00000036760
|
tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr2_-_2020044 | 11.42 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr17_-_4318393 | 11.31 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr20_+_20672163 | 11.22 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr2_+_59015878 | 11.20 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr25_-_25736958 | 10.67 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr8_-_4618653 | 10.45 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr7_+_23907692 | 10.01 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr9_-_48397702 | 9.72 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr9_-_296169 | 9.56 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr13_+_36622100 | 9.51 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr2_+_23222939 | 9.49 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr10_+_15777258 | 9.44 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr2_-_44282796 | 9.44 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr18_+_26895994 | 9.26 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr6_-_52156427 | 9.18 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr15_-_19250543 | 8.62 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr6_-_17849786 | 8.44 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr13_+_4405282 | 8.28 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr17_+_51764310 | 8.19 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr13_+_36764715 | 8.13 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr7_+_30867008 | 8.08 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr11_+_1796426 | 8.02 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr20_-_53366137 | 7.99 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr13_+_421231 | 7.95 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr5_-_69482891 | 7.87 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr15_+_47161917 | 7.87 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr17_-_43466317 | 7.54 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr10_+_37500234 | 7.51 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr9_+_34425736 | 7.31 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr13_-_30028103 | 7.24 |
ENSDART00000183889
|
scdb
|
stearoyl-CoA desaturase b |
| chr6_+_34512313 | 7.23 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr11_+_35364445 | 7.23 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr25_+_35375848 | 6.97 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr24_+_41931585 | 6.62 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr5_+_67812062 | 6.57 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr6_-_15604157 | 6.43 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr6_+_59967994 | 6.28 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr10_+_39952995 | 6.26 |
ENSDART00000183077
|
BX927333.1
|
|
| chr12_+_42436328 | 6.17 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr25_+_25737386 | 5.85 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr19_+_8144556 | 5.83 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr21_+_34167178 | 5.82 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr6_-_15604417 | 5.71 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr1_+_57371447 | 5.70 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr10_+_1638876 | 5.68 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr11_-_38083397 | 5.60 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr11_+_25064519 | 5.50 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr15_+_47418565 | 5.32 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr3_-_6767440 | 5.19 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr16_+_29303971 | 5.19 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr5_+_38263240 | 5.14 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr19_+_342094 | 5.10 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr6_-_18976168 | 5.10 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr21_+_1382078 | 4.97 |
ENSDART00000188463
|
TCF4
|
transcription factor 4 |
| chr25_-_11088839 | 4.95 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr20_-_36617313 | 4.95 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr7_-_18601206 | 4.83 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr17_+_35097024 | 4.75 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr11_-_4235811 | 4.71 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr2_+_25657958 | 4.55 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr24_-_21923930 | 4.54 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr22_-_881080 | 4.46 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr1_+_9004719 | 4.46 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr6_+_18520859 | 4.45 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr2_+_25658112 | 4.43 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr1_+_42874410 | 4.40 |
ENSDART00000153506
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr7_+_38762043 | 4.32 |
ENSDART00000036461
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr8_-_1698155 | 4.23 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr16_-_44399335 | 4.23 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr14_+_33264303 | 4.02 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr14_-_24391424 | 4.00 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr15_+_24388782 | 3.98 |
ENSDART00000191661
ENSDART00000179995 ENSDART00000111226 |
sez6b
|
seizure related 6 homolog b |
| chr19_-_34011340 | 3.92 |
ENSDART00000172618
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr12_+_11080776 | 3.92 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr8_+_20157798 | 3.88 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr3_-_16227490 | 3.79 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr16_+_53455638 | 3.75 |
ENSDART00000045792
ENSDART00000154189 |
rbm24b
|
RNA binding motif protein 24b |
| chr23_+_30730121 | 3.74 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr4_-_5018705 | 3.73 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr9_-_3149896 | 3.71 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr2_+_413370 | 3.71 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr20_+_35058634 | 3.70 |
ENSDART00000122696
|
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr17_+_8925232 | 3.62 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr24_-_12745222 | 3.54 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr4_-_5019113 | 3.49 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr23_-_32129569 | 3.47 |
ENSDART00000167761
ENSDART00000139569 |
zgc:92658
|
zgc:92658 |
| chr16_+_30002605 | 3.46 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr3_-_16227683 | 3.46 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr3_+_59117136 | 3.44 |
ENSDART00000182745
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr20_-_16156419 | 3.41 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr14_+_12109535 | 3.37 |
ENSDART00000054620
ENSDART00000121913 |
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr19_-_42391383 | 3.27 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr6_+_16031189 | 3.25 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr24_-_36238054 | 3.20 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr1_-_55058795 | 3.13 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr19_+_27479563 | 3.12 |
ENSDART00000049368
ENSDART00000185426 |
atat1
|
alpha tubulin acetyltransferase 1 |
| chr7_+_69528850 | 3.05 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr8_-_410199 | 3.04 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr5_+_61657702 | 3.03 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr23_-_41821825 | 3.01 |
ENSDART00000184725
|
GMEB2
|
si:ch73-302a13.2 |
| chr3_+_24537023 | 2.96 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr4_+_9011825 | 2.96 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr6_-_16406210 | 2.96 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr21_+_11415224 | 2.92 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr9_+_22375779 | 2.90 |
ENSDART00000183956
|
dgkg
|
diacylglycerol kinase, gamma |
| chr6_+_34511886 | 2.89 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr3_-_32362872 | 2.87 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr7_-_6592142 | 2.85 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr6_-_57938043 | 2.84 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr20_+_29565906 | 2.84 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr12_+_47909026 | 2.81 |
ENSDART00000192472
|
tbata
|
thymus, brain and testes associated |
| chr14_+_12110020 | 2.79 |
ENSDART00000192462
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr17_+_11507131 | 2.78 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr19_-_31686252 | 2.75 |
ENSDART00000131721
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr3_-_22829710 | 2.73 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr9_-_10145795 | 2.72 |
ENSDART00000004745
ENSDART00000143295 |
hnmt
|
histamine N-methyltransferase |
| chr17_-_44440832 | 2.71 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr25_-_20268027 | 2.65 |
ENSDART00000138763
|
dnajb9a
|
DnaJ (Hsp40) homolog, subfamily B, member 9a |
| chr22_-_6801876 | 2.64 |
ENSDART00000135726
|
si:ch1073-188e1.1
|
si:ch1073-188e1.1 |
| chr22_-_21676364 | 2.63 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr2_+_19236677 | 2.59 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr10_+_3299829 | 2.58 |
ENSDART00000183684
|
zgc:56235
|
zgc:56235 |
| chr21_-_39670375 | 2.54 |
ENSDART00000151567
|
sgk494b
|
uncharacterized serine/threonine-protein kinase SgK494b |
| chr4_+_9011448 | 2.52 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr22_+_34430310 | 2.52 |
ENSDART00000109860
|
amigo3
|
adhesion molecule with Ig-like domain 3 |
| chr11_-_42554290 | 2.50 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr3_+_5575313 | 2.50 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr7_-_59159253 | 2.48 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr21_+_10794914 | 2.48 |
ENSDART00000084035
|
znf532
|
zinc finger protein 532 |
| chr23_-_18707418 | 2.45 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr1_-_58036509 | 2.45 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr1_+_11977426 | 2.45 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr24_-_26632171 | 2.43 |
ENSDART00000008374
ENSDART00000017384 |
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr19_-_2421793 | 2.42 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr11_-_36341189 | 2.42 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr8_+_104114 | 2.41 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr3_+_31925067 | 2.40 |
ENSDART00000127330
ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr18_-_14917296 | 2.37 |
ENSDART00000098662
|
panx2
|
pannexin 2 |
| chr19_+_27479838 | 2.35 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr11_+_29790626 | 2.32 |
ENSDART00000067822
|
dynlt3
|
dynein, light chain, Tctex-type 3 |
| chr20_-_43786515 | 2.29 |
ENSDART00000004601
|
laptm4a
|
lysosomal protein transmembrane 4 alpha |
| chr10_+_5135842 | 2.28 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr24_-_35534273 | 2.27 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr11_-_36341028 | 2.23 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr5_+_61658282 | 2.21 |
ENSDART00000188878
|
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr23_-_44466257 | 2.18 |
ENSDART00000150126
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr22_-_2922053 | 2.16 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr8_-_17167819 | 2.15 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr10_+_6383270 | 2.13 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr9_+_4378153 | 2.11 |
ENSDART00000191264
ENSDART00000182384 |
kalrna
|
kalirin RhoGEF kinase a |
| chr2_+_30916188 | 2.10 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr12_+_49125510 | 2.10 |
ENSDART00000185804
|
FO704607.1
|
|
| chr1_+_45922699 | 2.09 |
ENSDART00000033669
|
lipt1
|
lipoyltransferase 1 |
| chr15_-_19128705 | 2.09 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr8_-_13046089 | 2.09 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr3_+_472158 | 2.08 |
ENSDART00000134971
|
zgc:194659
|
zgc:194659 |
| chr21_+_21279159 | 2.07 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr18_-_46258612 | 2.06 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr17_-_17759138 | 2.04 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr20_-_51656512 | 2.00 |
ENSDART00000129965
|
LO018154.1
|
|
| chr6_+_29693492 | 1.99 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr14_+_24277556 | 1.97 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr17_+_6765621 | 1.97 |
ENSDART00000156637
ENSDART00000007622 |
afg1la
|
AFG1 like ATPase a |
| chr1_+_45707219 | 1.96 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr10_-_42131408 | 1.96 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr21_-_22122312 | 1.94 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr16_-_20294236 | 1.94 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr10_+_4875262 | 1.92 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr11_-_11301283 | 1.89 |
ENSDART00000113311
ENSDART00000180466 |
col9a1a
|
collagen, type IX, alpha 1a |
| chr11_+_45287541 | 1.87 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr24_-_14712427 | 1.87 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr7_-_31938938 | 1.86 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr1_-_55196103 | 1.85 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr8_-_410728 | 1.83 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr13_-_6252498 | 1.81 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr9_-_7655243 | 1.81 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr14_-_47849216 | 1.80 |
ENSDART00000192796
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr10_+_41668483 | 1.78 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr2_+_37134281 | 1.77 |
ENSDART00000020135
|
pex19
|
peroxisomal biogenesis factor 19 |
| chr5_-_64431927 | 1.75 |
ENSDART00000158248
|
brd3b
|
bromodomain containing 3b |
| chr3_-_52899394 | 1.74 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr10_+_1396940 | 1.74 |
ENSDART00000150096
|
gdnfa
|
glial cell derived neurotrophic factor a |
| chr7_-_3470451 | 1.73 |
ENSDART00000173387
|
si:ch211-285c6.6
|
si:ch211-285c6.6 |
| chr16_+_12812472 | 1.72 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr14_-_24277805 | 1.70 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr5_-_2689753 | 1.67 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr17_+_16429826 | 1.66 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr23_-_27822920 | 1.66 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr2_+_19236969 | 1.65 |
ENSDART00000163875
ENSDART00000168644 |
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr3_+_36284986 | 1.65 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 2.7 | 10.7 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 2.4 | 9.6 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 1.8 | 7.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.7 | 5.2 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 1.6 | 4.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.1 | 4.4 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 1.1 | 6.6 | GO:0089718 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.9 | 3.7 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.9 | 3.7 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.9 | 2.8 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.9 | 2.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.8 | 4.0 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.7 | 6.7 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.7 | 2.9 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.7 | 7.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.7 | 2.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.7 | 13.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.6 | 3.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.6 | 13.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.6 | 4.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.6 | 1.7 | GO:0061213 | regulation of neurotransmitter uptake(GO:0051580) regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.5 | 1.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.5 | 7.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 5.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.5 | 7.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.5 | 1.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.5 | 7.0 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 1.8 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.4 | 2.8 | GO:0036268 | swimming(GO:0036268) |
| 0.4 | 2.4 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.4 | 4.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 13.2 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.3 | 2.4 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 3.2 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.3 | 10.1 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.3 | 0.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 2.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 1.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 | 7.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 1.9 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.3 | 3.0 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.3 | 10.0 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 8.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.2 | 2.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 16.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.2 | 1.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.2 | 5.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 9.3 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.2 | 1.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 1.9 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.8 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.2 | 1.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 1.6 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 3.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 5.1 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.2 | 3.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.7 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 1.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 15.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 2.5 | GO:0034661 | ncRNA catabolic process(GO:0034661) |
| 0.1 | 1.1 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.6 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 1.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 3.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.5 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 2.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 3.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.1 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 14.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 1.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 2.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 2.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 10.7 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.1 | 2.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.5 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 5.5 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.4 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 7.9 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 1.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 10.3 | GO:0044782 | cilium organization(GO:0044782) |
| 0.1 | 8.0 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.1 | 0.3 | GO:0061549 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) sympathetic ganglion development(GO:0061549) |
| 0.1 | 38.5 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.1 | 2.0 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 2.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.6 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 6.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 0.8 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.1 | 1.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.7 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 2.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 4.6 | GO:0018393 | internal protein amino acid acetylation(GO:0006475) internal peptidyl-lysine acetylation(GO:0018393) |
| 0.0 | 0.7 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 1.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 8.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 2.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 2.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.8 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 2.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 1.7 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 2.4 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.8 | GO:0042181 | ketone biosynthetic process(GO:0042181) |
| 0.0 | 4.7 | GO:0046578 | regulation of Ras protein signal transduction(GO:0046578) |
| 0.0 | 0.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 12.1 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 1.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 2.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 3.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.7 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 6.8 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 0.4 | GO:0006623 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 2.0 | 7.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 1.5 | 6.2 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 1.4 | 13.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 1.3 | 22.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 1.1 | 8.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 1.0 | 16.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.7 | 2.1 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.7 | 2.8 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.7 | 5.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.5 | 3.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.5 | 13.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.4 | 7.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 3.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 15.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.4 | 1.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.3 | 33.6 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.3 | 1.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.3 | 3.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.3 | 2.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 7.4 | GO:0005844 | polysome(GO:0005844) |
| 0.3 | 9.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 2.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 2.1 | GO:0044447 | axoneme part(GO:0044447) |
| 0.2 | 9.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 1.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 1.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 5.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 12.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 2.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 1.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 5.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 31.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 6.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 7.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 2.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 15.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 7.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 4.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 5.8 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 7.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 15.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.6 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 4.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.6 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.8 | 7.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.4 | 33.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 1.3 | 4.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 1.3 | 9.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.9 | 6.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.9 | 2.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.9 | 2.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.9 | 4.5 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.9 | 13.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.8 | 2.5 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.8 | 7.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.7 | 3.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.7 | 3.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.7 | 2.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.7 | 13.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.6 | 7.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 3.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.5 | 1.6 | GO:0034618 | arginine binding(GO:0034618) |
| 0.5 | 2.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.5 | 3.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 9.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 1.9 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.4 | 1.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 5.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 1.7 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.3 | 5.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 3.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 2.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.3 | 3.9 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.3 | 1.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.3 | 9.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 3.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 0.8 | GO:0000035 | acyl binding(GO:0000035) |
| 0.3 | 1.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 3.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 5.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 7.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 13.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 0.7 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 0.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 2.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 10.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 0.8 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 3.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 2.9 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.2 | 4.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 1.8 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 3.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 10.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.2 | 1.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 5.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 7.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.9 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 8.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 2.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 6.7 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.1 | 2.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 1.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 11.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 1.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.8 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 9.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.5 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 28.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 2.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 3.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 2.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 4.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 4.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 4.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 5.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.6 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 4.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 1.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 1.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.2 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 1.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 4.1 | GO:0016887 | ATPase activity(GO:0016887) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.4 | 3.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 8.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 11.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 7.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 8.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 4.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 5.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.8 | 8.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.8 | 10.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.5 | 5.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 4.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 6.8 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.3 | 3.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 3.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 7.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 1.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 5.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 2.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |