Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
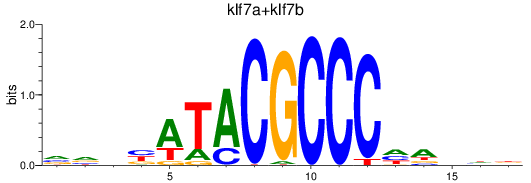
Results for klf7a+klf7b
Z-value: 0.34
Transcription factors associated with klf7a+klf7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf7b
|
ENSDARG00000043821 | Kruppel-like factor 7b |
|
klf7a
|
ENSDARG00000073857 | Kruppel-like factor 7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf7b | dr11_v1_chr9_-_28399071_28399071 | 0.58 | 1.3e-09 | Click! |
| klf7a | dr11_v1_chr1_-_6494384_6494384 | 0.46 | 3.7e-06 | Click! |
Activity profile of klf7a+klf7b motif
Sorted Z-values of klf7a+klf7b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_17441734 | 2.26 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr23_-_12015139 | 1.95 |
ENSDART00000110627
ENSDART00000193988 ENSDART00000184528 |
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr2_-_14390627 | 1.95 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr19_+_23982466 | 1.85 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr16_+_53519048 | 1.82 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr7_-_2039060 | 1.65 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr23_-_12014931 | 1.49 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr5_-_35252761 | 1.26 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr19_+_31404686 | 1.25 |
ENSDART00000078459
|
pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr16_+_33163858 | 1.22 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr5_+_32791245 | 1.00 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr8_-_22288258 | 0.98 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr19_-_30422239 | 0.96 |
ENSDART00000043554
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr19_-_30422035 | 0.96 |
ENSDART00000144265
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr19_+_43669122 | 0.87 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr1_+_14073891 | 0.86 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr8_+_44475793 | 0.85 |
ENSDART00000190118
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr6_+_30668098 | 0.84 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr8_+_54013199 | 0.83 |
ENSDART00000158497
|
CABZ01079663.1
|
|
| chr24_-_21487609 | 0.82 |
ENSDART00000191325
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr20_+_20731633 | 0.77 |
ENSDART00000191952
ENSDART00000165224 |
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr2_+_54844755 | 0.76 |
ENSDART00000170428
|
rnf126
|
ring finger protein 126 |
| chr21_+_8427059 | 0.73 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr22_+_980290 | 0.73 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr1_+_16574312 | 0.68 |
ENSDART00000187067
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr20_+_20731052 | 0.67 |
ENSDART00000047662
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr18_-_6943577 | 0.67 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr17_+_43843536 | 0.65 |
ENSDART00000164097
|
FO704777.1
|
|
| chr11_-_16115804 | 0.65 |
ENSDART00000143436
ENSDART00000157928 |
rpf1
|
ribosome production factor 1 homolog |
| chr15_+_5132439 | 0.64 |
ENSDART00000010350
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr11_-_16215470 | 0.63 |
ENSDART00000185744
|
rab7
|
RAB7, member RAS oncogene family |
| chr2_-_42035250 | 0.61 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr1_+_39990547 | 0.61 |
ENSDART00000112150
|
sod3b
|
superoxide dismutase 3, extracellular b |
| chr17_-_37795865 | 0.61 |
ENSDART00000025853
|
zfp36l1a
|
zinc finger protein 36, C3H type-like 1a |
| chr7_-_30087048 | 0.59 |
ENSDART00000112743
|
nmbb
|
neuromedin Bb |
| chr10_-_41157135 | 0.57 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr1_+_26012668 | 0.51 |
ENSDART00000182428
|
BX323448.1
|
|
| chr3_-_28872756 | 0.49 |
ENSDART00000138840
|
eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr19_+_19511394 | 0.46 |
ENSDART00000172410
|
jazf1a
|
JAZF zinc finger 1a |
| chr7_+_24081167 | 0.46 |
ENSDART00000141749
|
acin1b
|
apoptotic chromatin condensation inducer 1b |
| chr16_-_13921589 | 0.43 |
ENSDART00000023543
|
rcvrn2
|
recoverin 2 |
| chr21_-_20120543 | 0.41 |
ENSDART00000065664
|
dusp4
|
dual specificity phosphatase 4 |
| chr5_-_26795438 | 0.38 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr24_+_22021675 | 0.38 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr25_+_35683956 | 0.37 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr11_-_2594045 | 0.35 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr15_-_12229874 | 0.29 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr13_+_40334727 | 0.25 |
ENSDART00000074950
|
slc25a28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr20_+_18741089 | 0.25 |
ENSDART00000183218
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr5_+_15202495 | 0.19 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr2_-_23172708 | 0.19 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr18_-_26894732 | 0.17 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr8_-_45759137 | 0.13 |
ENSDART00000123878
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr7_-_37563883 | 0.06 |
ENSDART00000148805
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr6_-_10034145 | 0.01 |
ENSDART00000185999
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf7a+klf7b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 0.6 | GO:0000303 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 1.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 1.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.0 | 0.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.3 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 1.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.6 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 1.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 3.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |